This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Snapshots of the smallest programmable nuclease TnpB published
A team led by Professor Virginijus Šikšnys from Vilnius University Life Sciences Center (VU LSC) determined the structure of TnpB using cryo-electron microscopy in collaboration with the group of Professor Guillermo Montoya at the Novo Nordisk Foundation Center for Protein Research (CPR) at the University of Copenhagen.
The article "TnpB structure reveals the minimal functional core of Cas12 nuclease family" was published in Nature.
CRISPR-Cas nucleases, such as Cas9 or Cas12, also known as gene scissors, have revolutionized the field of genome editing. They enable precise editing of genomes and correction of disease-causing mutations. However, the size of Cas9 or Cas12 limits their delivery to target cells using Adeno-associated viruses (AAV), which are already used in gene therapy.
In their previous Nature paper, VU LSC scientists reported the discovery of a new class of programmable nucleases called TnpBs, which are associated with mobile genetic elements called transposons. They demonstrated that TnpB is the smallest programmable nuclease that can be applied for efficient gene editing; however, its structural organization and mechanism remained unknown.
"The new Nature paper that just came out is a result of a consistent and long-term effort that demonstrates the potential of Lithuanian scientists in the field of life sciences and their ability to be among the leaders in this field. This research has revealed the structure and mechanism of TnpB gene scissors, which creates a basis for further targeted engineering of the TnpB complex to transform it into a therapeutic tool for treating genetic disease," says Professor V. Šikšnys.
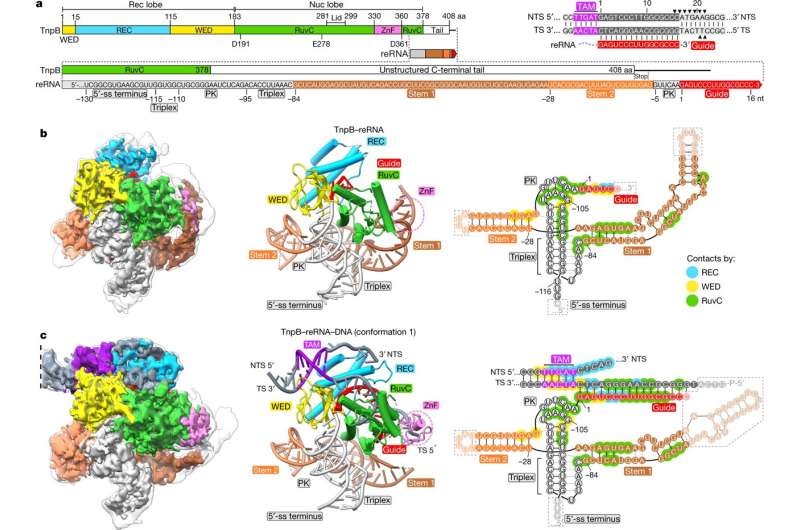
In the current study, scientists used cryo-electron microscopy (cryo-EM) to determine the ternary structures of the smallest programmable endonuclease, TnpB, which, along with biochemical studies, explained how TnpB gene scissors can precisely recognize and cut DNA targets.
Structural studies revealed that a long RNA molecule associated with the TnpB protein forms a complex three-dimensional structure that not only helps to recognize the DNA target but also controls TnpB's DNA-cutting activity. A comparison of structures and bioinformatic analysis revealed that TnpB is the precursor of the Cas12 nuclease family and forms the Cas12 structural-functional core.
As noted by one of the article's authors, Dr. Giedrius Sasnauskas, the success of this research was determined by several factors. "First of all—the relevant research object and the collaboration of VU LSC biochemists, molecular biologists, bioinformaticians, and colleagues from NNF-CPR at the University of Copenhagen. But most importantly, we were able to conduct this research in Lithuania using the cryo-electron microscope available at VU LSC," says the researcher.
More information: Giedrius Sasnauskas et al, TnpB structure reveals minimal functional core of Cas12 nuclease family, Nature (2023). DOI: 10.1038/s41586-023-05826-x
Journal information: Nature
Provided by Vilnius University