This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New digital tool could change the way we see cells
Being able to observe and track the way cells change and develop over time is a vital part of scientific and medical research.
Time-lapse studies of cells can show us how cells have mutated in certain environments or their reactions to external influences, such as medical treatment. This information can shine a light on how disease spreads and why some patients' cells do not respond to treatment such as chemotherapy.
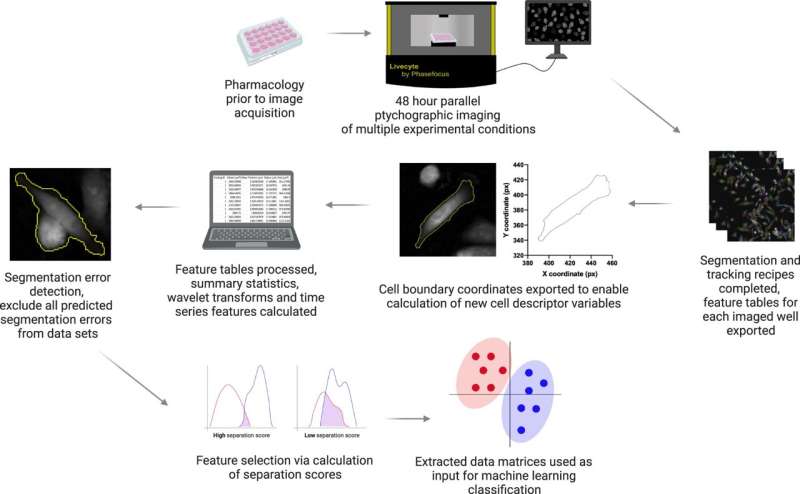
Tracking the development of specific cell features is a difficult process however, scientists at the departments of Biology and Mathematics at the University of York have now created a free digital tool which can help.
The software package, called CellPhe, is the first of its kind as it can extract a series of features from a cell in a time-lapse study and characterize the cells based on their behavior and internal structure. The package also automatically removes errors in cell tracking to improve data quality.
In a study published April 3 in Nature Communications, CellPhe correctly identified two different sets of breast cancer cells—one which was treated with chemotherapy drugs and the other which was not. Comparing the two groups, CellPhe was also able to identify a potentially resistant subset of treated cells.
Data like this is particularly important in understanding breast cancer and designing its treatment, as chemoresistance commonly leads to relapse in breast cancer patients.
CellPhe could also have further practical applications in processes such as drug screening and the prediction of disease prognosis.
Laura Wiggins from the Department of Biology said, "It is hugely exciting to be able to quantify cell behavior over time in such unprecedented detail.
"A lot of hard work has gone into making CellPhe user-friendly and adaptable to new applications so we look forward to seeing how our toolkit will be used by the community.
We foresee CellPhe playing a pivotal role in our understanding of cellular drug response as well as the ways in which cells communicate with one another, via signaling as well as through direct contact."
CellPhe is freely available online and will run on any operating system and comes with a manual and an instruction video.
More information: Laura Wiggins et al, The CellPhe toolkit for cell phenotyping using time-lapse imaging and pattern recognition, Nature Communications (2023). DOI: 10.1038/s41467-023-37447-3
Journal information: Nature Communications
Provided by University of York