This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers develop a new way to identify bacteria in fluids
Shine a laser on a drop of blood, mucus, or wastewater, and the light reflecting back can be used to positively identify bacteria in the sample.
"We can find out not just that bacteria are present, but specifically which bacteria are in the sample—E. coli, Staphylococcus, Streptococcus, Salmonella, anthrax, and more," said Jennifer Dionne, an associate professor of materials science and engineering and, by courtesy, of radiology at Stanford University. "Every microbe has its own unique optical fingerprint. It's like the genetic and proteomic code scribbled in light."
Dionne is senior author of a new study in the journal Nano Letters detailing an innovative method her team has developed that could lead to faster (almost immediate), inexpensive, and more accurate microbial assays of virtually any fluid one might want to test for microbes.
Traditional culturing methods still in use today can take hours if not days to complete. A tuberculosis culture takes 40 days, Dionne said. The new test can be done in minutes and holds the promise of better and faster diagnoses of infection, improved use of antibiotics, safer foods, enhanced environmental monitoring, and faster drug development, says the team.
Old dogs, new tricks
The breakthrough is not that bacteria display these spectral fingerprints, a fact that has been known for decades, but in how the team has been able to reveal those spectra amid the blinding array of light reflecting from each sample.
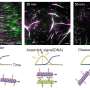
"Not only does each type of bacterium demonstrate unique patterns of light but virtually every other molecule or cell in a given sample does too," said first author Fareeha Safir, a Ph.D. student in Dionne's lab. "Red blood cells, white blood cells, and other components in the sample are sending back their own signals, making it hard if not impossible to distinguish the microbial patterns from the noise of other cells."
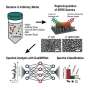
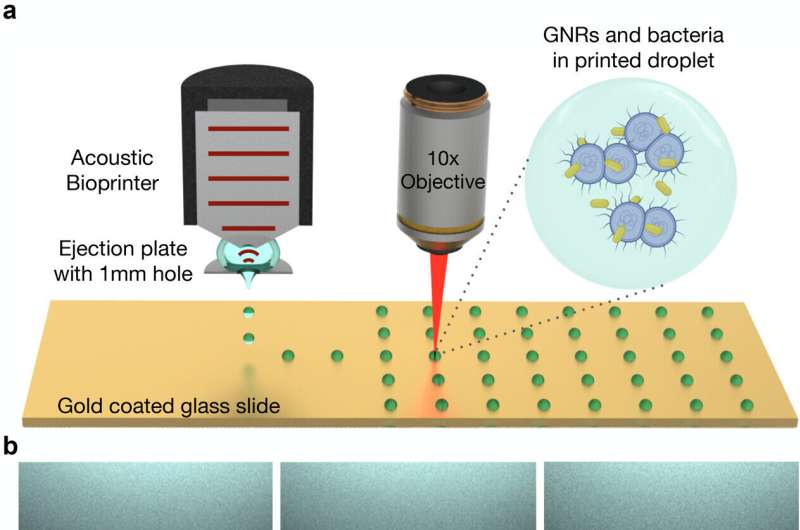
A milliliter of blood—about the size of a raindrop—can contain billions of cells, only a few of which might be microbes. The team had to find a way to separate and amplify the light reflecting from the bacteria alone. To do that, they ventured along several surprising scientific tangents, combining a four-decade-old technology borrowed from computing—the inkjet printer—and two cutting-edge technologies of our time—nanoparticles and artificial intelligence.
"The key to separating bacterial spectra from other signals is to isolate the cells in extremely small samples. We use the principles of inkjet printing to print thousands of tiny dots of blood instead of interrogating a single large sample," explained co-author Butrus "Pierre" Khuri-Yakub, a professor emeritus of electrical engineering at Stanford who helped develop the original inkjet printer in the 1980s.
"But you can't just get an off-the-shelf inkjet printer and add blood or wastewater," Safir emphasized. To circumvent challenges in handling biological samples, the researchers modified the printer to put samples to paper using acoustic pulses. Each dot of printed blood is then just two trillionths of a liter in volume—more than a billion times smaller than a raindrop. At that scale, the droplets are so small they may hold just a few dozen cells.
In addition, the researchers infused the samples with gold nanorods that attach themselves to bacteria, if present, and act like antennas, drawing the laser light toward the bacteria and amplifying the signal some 1500 times its unenhanced strength. Appropriately isolated and amplified, the bacterial spectra stick out like scientific sore thumbs.
The final piece of the puzzle is the use of machine learning to compare the several spectra reflecting from each printed dot of fluid to spot the telltale signatures of any bacteria in the sample.
"It's an innovative solution with the potential for life-saving impact. We are now excited for commercialization opportunities that can help redefine the standard of bacterial detection and single-cell characterization," said senior co-author Amr Saleh, a former postdoctoral scholar in Dionne's lab and now a professor at Cairo University.
While this technique was created and perfected using samples of blood, Dionne is equally confident that it can be applied to other sorts of fluids and target cells beyond bacteria, like testing drinking water for purity or perhaps spotting viruses faster, more accurately, and at lower cost than present methods.
More information: Fareeha Safir et al, Combining Acoustic Bioprinting with AI-Assisted Raman Spectroscopy for High-Throughput Identification of Bacteria in Blood, Nano Letters (2023). DOI: 10.1021/acs.nanolett.2c03015
Journal information: Nano Letters
Provided by Stanford University