This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Research team develops new protein mapping technique
A groundbreaking technique for in-depth mapping of protein localizations in whole tissue developed by researchers in the University at Buffalo School of Pharmacy and Pharmaceutical Sciences has been published in Nature Communications.
Most biological functions, drug effects and diseases occur in tissues, which are often considered a homogenous entity in which different types of cells are woven in regularly repeated patterns. However, a tissue is more like a city, with highly diversified, dynamic features across different regions.
When trying to understand a city, one does not study it as a singularity, but instead investigate its activities and functions at different locations. Because of the complex interactions within tissues, it is critically important to investigate the location-specific biological activities across all tissue regions.
Increasing evidence suggests that the information on spatially resolved biological regulations, which are missed in the majority of current investigations, holds the key to addressing intractable diseases and to discovering and developing novel drugs.
MASP technique maps thousands of proteins on whole tissue
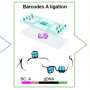
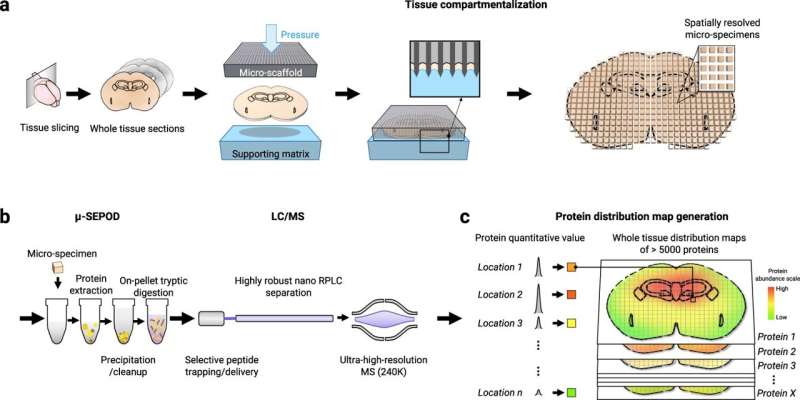
Spatially resolved analysis on a proteome level is highly valuable in pharmaceutical and clinical investigations, but is challenging. Over the past three years, Jun Qu and colleagues—among them co-first authors Min Ma, a doctoral student at Roswell Park Comprehensive Cancer Center; Shihan Huo, postdoctoral associate in the Department of Pharmaceutical Sciences; Ming Zhang, a research scientist in the Department of Pharmaceutical Sciences; and Shuo Qian, a doctoral student at Roswell Park—developed Micro-scaffold Assisted Spatial Proteomic (MASP).
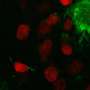
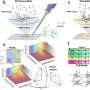
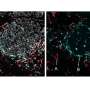
For the first time, this technique has enabled the mapping of thousands of proteins on a whole tissue with excellent accuracy and precision. One of the key innovations is a revolutionary, micro-scaffold tissue compartmentalization device fabricated using state-of-the-art, 3D printing techniques. This method is capable of simultaneously mapping numerous proteins across a whole tissue slice in a single experiment, which had never been achieved before. The teamed showed an application of the MASP pipeline by mapping more than 5,000 proteins with high accuracy in a healthy mouse brain. It included many important brain markers, regulators and transporters, where most of the proteins were for the first time mapped on the whole-tissue level.
"The uneven distribution of biotherapeutics in tissues is frequently responsible for compromised efficacy and undesirable side effects in clinical settings," Qu explains. "We believe that understanding the spatial distribution of biotherapeutics on a whole-tissue level will afford critical information for directing pharmaceutical efforts in evaluating and designing more effective and safer biotherapeutics. MASP has made this happen by allowing a simultaneous study of the intra-brain distribution of the protein drug but also the spatially related biomarkers."
Unique protein mapping in signaling networks, other pathways
One unique advantage of MASP is that it allows mapping of many important proteins in various signaling networks, researchers say. For example, MASP generated whole-tissue distribution maps for proteins involved in the signaling pathway of Alzheimer's disease. For most proteins in the pathway, this was the first time their cerebral protein distributions have been observed. The research also showed the first-ever mapping of key players in other disease and function pathways.
"Compared to previous methods for studying protein spatial distribution, such as immunoassays, MASP faithfully preserves the spatial information based on a robust compartmentalization of the tissue, which does not introduce any artifacts related to the labeling process," Ma and Huo note. "More importantly, MASP accurately quantifies each protein at each location on the tissue, which is not affected by the limited dynamic range observed in immunoassays."
The second generation of MASP, with higher resolution and higher throughput, is currently being developed in Qu's lab. The team anticipates a wide range of applications for this novel strategy, such as understanding the efficacy/safety issues of drugs and investigation of the onset and development of diseases.
More information: Min Ma et al, In-depth mapping of protein localizations in whole tissue by micro-scaffold assisted spatial proteomics (MASP), Nature Communications (2022). DOI: 10.1038/s41467-022-35367-2
Journal information: Nature Communications
Provided by University at Buffalo