This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers propose rapid identification and drug resistance screening of respiratory pathogens
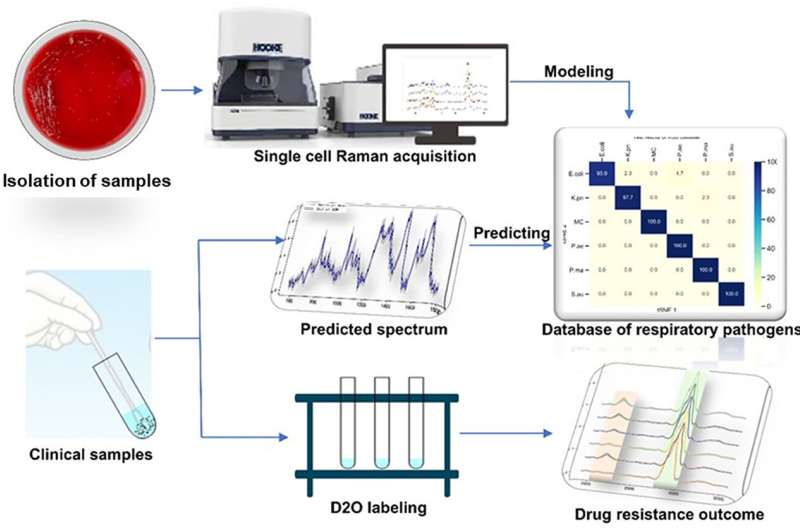
In a study published in Frontiers in Microbiology, a cooperative research group from the Changchun Institute of Optics, Fine Mechanics and Physics (CIOMP) of the Chinese Academy of Sciences (CAS) and Jilin University proposed rapid identification and drug resistance screening of pathogenic bacteria based on single cell Raman spectroscopy (SCRS).
SCRS is a "whole biological fingerprint" technique that can be used to identify microbial samples. It can be used for in situ, non-invasive, and non-labeled detection of samples and has a great application value for qualitative analysis, quantitative analysis, and molecular structure determination.
The combination of D2O isotope labeling technology and SCRS can identify the Raman shift caused by the difference of metabolic activity between cells, which can be used as a semi-quantitative method to identify the metabolic activity of cells. The combination of the Raman and heavy water labeling techniques at the single-cell level can overcome the requirement of long-term culture in clinical pathogen experiments, making rapid drug screening possible.
In this study, a SCRS technique combined with a heavy water labeling was designed as a rapid and accurate method for detecting antibiotic-resistant bacteria in respiratory bacteria. And a Raman phenotype database of six common respiratory pathogens was established to provide a new method for rapidly identifying respiratory pathogens in clinical practice. Different antibiotic-heavy water labeling conditions were also explored to achieve drug resistance identification of respiratory pathogens within two hours.
Results showed the classification accuracy of the isolated samples was 93–100%, and the classification accuracy of the clinical samples was more than 80%. Combined with heavy water labeling technology, the drug resistance of respiratory tract pathogens was determined.
The study showed that SCRS–D2O labeling could rapidly identify drug resistance of respiratory tract pathogens within two hours, which provides a rapid and sensitive method for identifying respiratory tract infection bacteria and their drug susceptibility.
More information: Ziyu Liu et al, Rapid identification and drug resistance screening of respiratory pathogens based on single-cell Raman spectroscopy, Frontiers in Microbiology (2023). DOI: 10.3389/fmicb.2023.1065173
Journal information: Frontiers in Microbiology
Provided by Chinese Academy of Sciences


















