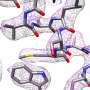
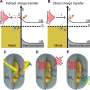
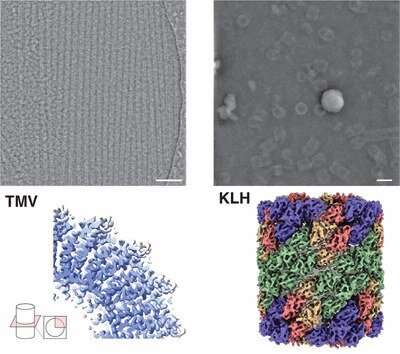
A new tool for cryo-electron microscopy
Researchers at Forschungszentrum Jülich and Heinrich Heine University Düsseldorf led by Prof. Dr. Carsten Sachse are using cryo-electron microscopy, or cryo-EM for short, to make biomolecules visible at the atomic level. In a paper now published in the journal Nature Methods, they present a new method that combines cryo-EM with a method otherwise used in materials research. The results are also presented and classified in a Nature Briefing.
The still relatively new technique of cryo-EM has a decisive advantage over X-ray crystallography that has been routinely in use for decades: Protein building blocks can be observed in their natural environment in a snap-frozen state without having to convert them into an artificial crystal beforehand. Cryo-EM is based on transmission electron microscopy. The alternative method that the researchers have now employed, on the other hand, is a further development of scanning transmission electron microscopy with integrated differential phase contrast, or iDPC-STEM for short.
"This method has so far been used primarily in materials research, where it has already led to very high resolutions. When imaging biological samples, we have now directly achieved a quality that was first made possible by cryo-electron microscopy a few years ago," says Carsten Sachse, Director at the Ernst Ruska-Centre of Forschungszentrum Jülich and Professor at Heinrich Heine University Düsseldorf.
Together with research partners from the analytics company Thermo Fisher Scientific in Eindhoven, he was able to map protein structures using iDPC-STEM with a sub-nanometer resolution of 3.5 angstroms. "Cryo-electron microscopy is a bit more advanced today in comparison. But our results show that iDPC-STEM is in principle capable, with some optimization, of achieving similar resolutions to today's cryo-EM and expanding the possibilities for structural analysis; especially for very heterogeneous, non-uniform samples or single particles when averaging capabilities are limited," says Carsten Sachse.

In conventional cryo-electron microscopy, thousands, sometimes tens or hundreds of thousands, of snapshots of a sample are taken from many viewing directions. A powerful computer uses these images to calculate a detailed three-dimensional model of the molecule or particle. Scanning electron microscopy, on the other hand, scans objects line by line in tiny steps to produce a composite image that, as in conventional cryo-EM, serves as the basis for the three-dimensional structure calculation. As with cryo-electron microscopy, a low dose electron beam is used because biomolecules are typically extremely sensitive. This prevents the high energy of the beam from destroying the sensitive structures.
More information: Ivan Lazić et al, Single-particle cryo-EM structures from iDPC–STEM at near-atomic resolution, Nature Methods (2022). DOI: 10.1038/s41592-022-01586-0
Obtaining cryo-EM structures by scanning transmission electron microscopy, Nature Methods (2022). DOI: 10.1038/s41592-022-01587-z
Journal information: Nature Methods , Nature
Provided by Forschungszentrum Juelich