Team develops method to increase gene editing efficiency while minimizing DNA deletion sizes
Wake Forest Institute for Regenerative Medicine (WFIRM) scientists working on CRISPR/Cas9-mediated gene editing technology have developed a method to increase efficiency of editing while minimizing DNA deletion sizes, a key step toward developing gene editing therapies to treat genetic diseases.
CRISPR (clustered regularly interspaced short palindromic repeats) technology is used to alter DNA sequences and modify gene function. CRISPR/Cas9 is an enzyme that is used like a pair of scissors to cut two strands of DNA at a specific location to add, remove or repair bits of DNA. By modifying gene function, scientists hope to treat genetic diseases by halting a diseased cell's ability to continue replicating the defective DNA. CRISPR/Cas9 is the most versatile genetic manipulation available and has a wide range of potential applications. While CRISPR/Cas9 mainly generates short insertions or deletions at the target site, it may also make large DNA deletions around the specific target site. These large deletions cause safety concerns and may decrease functional editing efficiency.
The WFIRM team is looking for ways to reduce the chances of this happening. The research described in their recent paper, published recently in Nucleic Acids Research, sought to address the generation of unpredictable on-target long DNA deletions and find a way to guard against them, said lead author Baisong Lu, Ph.D., of WFIRM.
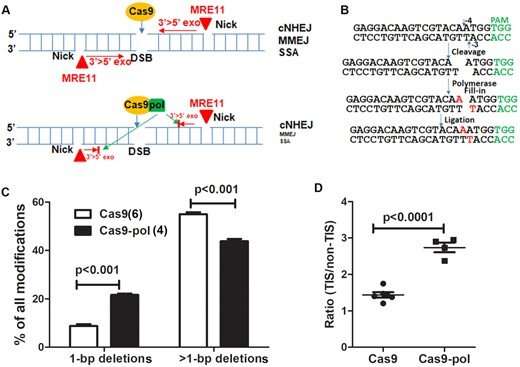
"Considering that a lack of methods to avoid generating long deletions by CRISPR/Cas9 is a challenge to the field, we are developing new counter strategies," Lu said. "We developed a method to refine the mutations—to increase small deletions and decrease large deletions." The team evaluated a variety of human cells and genes of interest and found that fusing DNA polymerase I or the Klenow fragment to the Cas9 enzyme minimized unforeseen large genomic DNA deletions without sacrificing genome editing efficiency. On the contrary, doing so even increased editing efficiency in human primary cells.
"This technique increased genome editing efficiency in primary cells and did not increase DNA substitution rates or off-target rates," said WFIRM Director Anthony Atala, MD, a co-author of the paper. "It also decreased large deletions, thus increasing safety. We can increase the percentage of desirable types of mutations. This improves the efficiency of disrupting disease-causing genes or restoring disrupted genes."
Co-authors include Kyung W. Yoo and Manish Kumar Yadav of WFIRM, and Qianqian Song of the Department of Cancer Biology, Wake Forest University Health Sciences.
More information: Kyung W Yoo et al, Targeting DNA polymerase to DNA double-strand breaks reduces DNA deletion size and increases templated insertions generated by CRISPR/Cas9, Nucleic Acids Research (2022). DOI: 10.1093/nar/gkac186
Journal information: Nucleic Acids Research
Provided by Wake Forest University Baptist Medical Center