May 4, 2022 feature
Reactive microscopy with MicroMator software
Microscopic imaging analysis is a crucial component of biochemistry and medicine, with significant progress in accuracy and speed made due to machine learning methods and improved computation. These technical advances can assist researchers adapt microscopy experimental plans online for real-time information of experimental observations. In a new study now published in Nature Communications, Zachary R. Fox and a team of interdisciplinary scientists in France and the U.S., developed MicroMator—an open and flexible software integrated with Python to drive reactive microscopy experiments. Using the experimental framework, they performed dynamic adaptation of fluorescence microscopy on bacteria to show the potential of MicroMator for real-time tracking and targeting experiments at the single-cell level in two primary case studies.
Software for reactive microscopy
Microscopy automation can be regulated with smart software to support reproducible high throughput microscopy experiments. However, many such experiments cannot be altered via human or computer-driven interventions, while the experiment is ongoing. For instance, during a reactive experiment, a researcher may need to trigger autofocus during the loss of focus, detect rare events and adapt the imaging routine or even send an email to highlight the detection of interesting anomalies that necessitate unplanned human intervention. While imaging analysis technology has made great leaps with deep learning software, the full-scope of microscopy automation remains to be explored. In this work, Fox et al presented MicroMator, a software that supports reactive microscopy experiments and explored two case studies as proof-of-concept.
Implementing MicroMator software
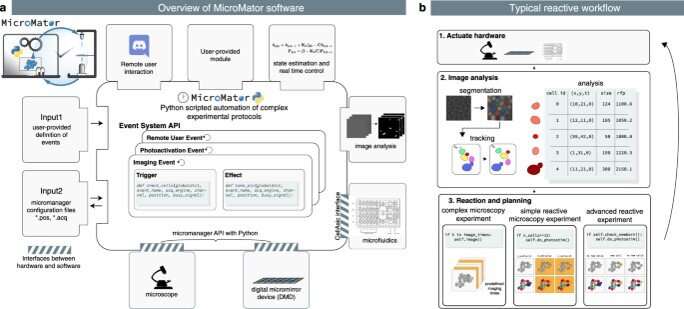
During the experiments, the team used MicroMator to define microscopic analyses via a main image acquisition loop that served as a backbone for experiments, to create functions and implement reactivity. As a result, the team modified the main acquisition loop relative to event effects during the course of the experiment. They designed the software using Python 3 an open-source algorithm with a modular design, where MicroMator provided a command line software to read configuration files defining the reactive experiments. MicroMator also included Micro-Manager generated files to specify the positions of interest during the experiment. The team created events after each image acquisition to conceive highly complex experiments, they required Python programming skills initially to develop the software, for debugging maintenance and reuse. During the study, Fox et al designed experiments to regulate cellular processes in real-time, inspired by previous studies.
-
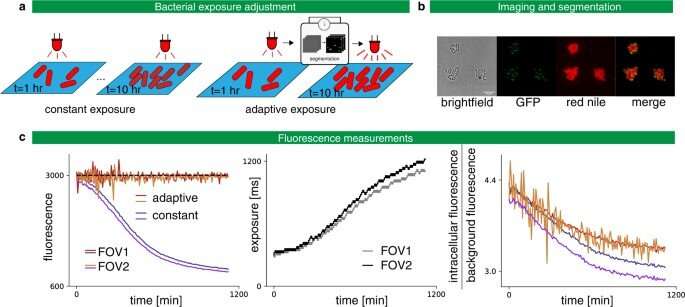
Adaptive exposure preserves imaging quality. a Left: Classic constant exposure experiment, in which the same amount of light is sent throughout the experiment. Right: adaptive experiment, in which bacteria are segmented after each frame and the exposure is increased or decreased depending on the measured fluorescence. b Imaging C. glutamicum in brightfield and fluorescence with Wag31 fused to mNeonGreen and localized to the poles (GFP), and with red nile stain. c Left: In the constant exposure experiment, the fluorescence decays. For the adaptive experiment, fluorescence tracks the target value of 3000 arb. units. The experiment is performed in two fields of view (FOV) in parallel. Center: The exposure settings as a function of experiment time. Right: Signal-to-noise ratio as defined by the intracellular fluorescence divided by the background fluorescence as a function of the experiment time. A less severe degradation is obtained with the adaptative strategy. Credit: Nature Communications (2022). DOI: 10.1038/s41467-022-29888-z -
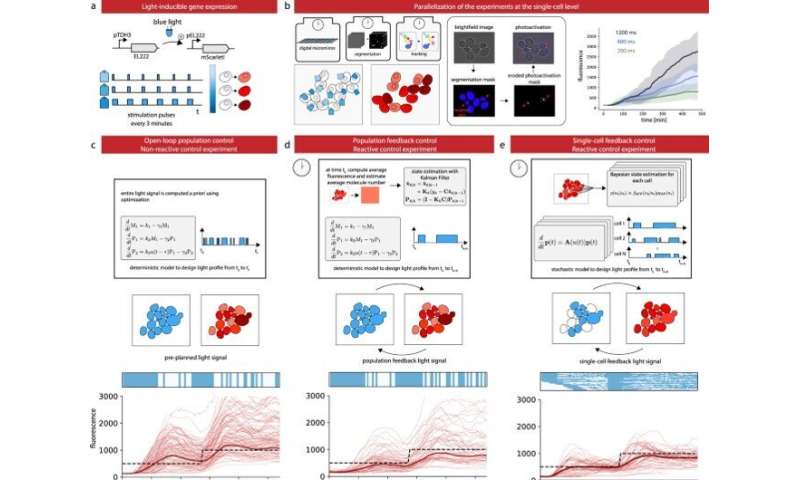
Control gene expression at the single cell level in yeast. a The red fluorescent protein mScarletI is placed under the control of the light-responsive transcription factor EL222. b To efficiently characterize cell responses to light stimulations, cells in the field of view are partitioned in three groups, each group being stimulated with a different temporal profile. Bright-field images are segmented and cells are tracked. Then, based on their groups, cells are stimulated during the appropriate time with eroded masks (see Supplementary Fig. 5). Therefore, characterization experiments are run in parallel thanks to the DMD and our capacity to segment and track cells in real time. The temporal evolution of the mean mScarletI fluorescence of the cells in the three groups is shown with envelopes indicating one standard deviation. Single-cell trajectories and replicates are provided in Supplementary Fig. 6. c Open-loop control experiment in which a model of the response of the cell population is used to precompute a light stimulation profile that drives the cell population to the target behavior. The application of the light profile leads to significant deviations from the target of the individual cell trajectories. d Closed-loop control experiment in which the same model is used jointly with real-time observations of the population state to decide which light profile to apply to all cells, using a receding horizon strategy. e A stochastic model of individual cell response is used jointly with single-cell observations to decide which light profile to apply to each cell. f The different strategies have similar performances to drive the mean fluorescence to its target, but the single-cell feedback strategy is significantly better to drive individual cells to their target profiles. Box plots indicate the lower quartile, the median, and the upper quartile of the target error, with the whiskers corresponding to 1.5 interquartile ranges. Each control experiment was replicated two times. Credit: Nature Communications (2022). DOI: 10.1038/s41467-022-29888-z
Case studies: Real-time experiments in bacteria
Using the experimental setup, Fox et al studied the Corynebacterium glutamicum strains, a model organism to understand Mycobacterium tuberculosis. After growing the cells of interest on agar pads, the cells expressed cell-cycle marker Wag31 fused to the fluorescent protein mNeonGreen and the Nile red lipolytic dye of the cell membrane.
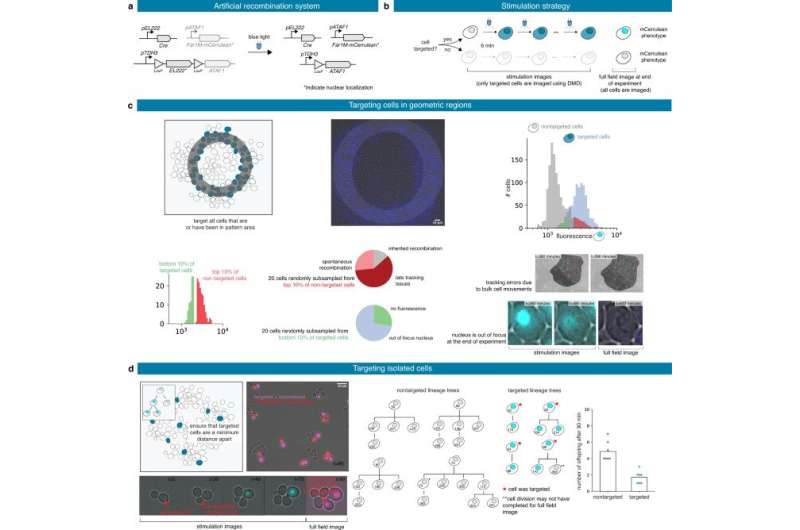
Typically, the complexity of the experiment can be monitored via different acquisition settings for different timeframes, however, in this instance, to overcome such challenges, Fox et al included MicroMator to achieve targeted fluorescence values for the red and green channels, and acquired a number of positions and their respective exposure during the experiments. The team then measured the intracellular fluorescence to perform time-sensitive bacteria segmentation via Cellpose—a deep learning-based generalist algorithm. In the second case study, the team used an optogenetic system and the mScarlet1 fluorescence reporter to build light-responsive yeast cells. For this, the team stimulated different cells in the field of view via a digital micromirror device with real-time imaging segmentation, and cell tracking features to regulate protein expression levels in a cell population. Fox et al used SegMator image analysis available in the setup, to segment and track cells in the field of view in real-time and estimated the time of fluorescence of reporter detection to account for the lag between cell stimulation and fluorescence detection during the experiment. In the third case study, Fox et al constructed a light-driven artificial recombination system in yeast and employed different light stimulation strategies to acquire various structures of recombined cells. The work showed how some cells were erroneously targeted for recombination and verified the necessity of reactive software for real-time image analysis at single-cell resolution.
Outlook
In this way, Zachary R. Fox and colleagues presented the MicroMator software to assist automated microscopy platforms reach their fullest potential. The platform can be used to conduct highly complex experiments, however, the team expect to primarily automate classical experiments, to highlight reactiveness in microscopy. The MicroMator software is relatively simple and applicable across most quantitative biology labs worldwide.
More information: Zachary R. Fox et al, Enabling reactive microscopy with MicroMator, Nature Communications (2022). DOI: 10.1038/s41467-022-29888-z
Michael Eisenstein, Smart solutions for automated imaging, Nature Methods (2020). DOI: 10.1038/s41592-020-00988-2
Journal information: Nature Communications , Nature Methods
© 2022 Science X Network