Three chaperones coordinate the breakup of amyloid fibrils in yeast
In a finding that has important ramifications for neurodegenerative disorders such as Alzheimer's disease, RIKEN researchers have discovered how three chaperone molecules in yeast cells work together to break up beta-sheet-rich fibrillar protein aggregates known as amyloid fibrils.
Proteins that are folded in the wrong way can't do their jobs properly. Some misfolded proteins known as prions are particularly insidious because they pass on their distorted shapes to the proteins around them, which then mesh together to form robust rafts called amyloid fibrils. Amyloid fibrils have been implicated in prion diseases (such as Creutzfeldt-Jakob disease), which are always fatal, and neurodegenerative disorders such as Alzheimer's and Parkinson's diseases.
The process by which amyloid fibrils form has received a lot of attention, but much less is known about the mechanism by which they are broken up in cells. This disaggregation process is important because the fragments it produces could allow prions to spread between cells.
"Many researchers have investigated the formation mechanism of amyloid fibrils, but there have been very few studies of amyloid disaggregation," says Motomasa Tanaka of the RIKEN Center for Brain Science. "However, the small protein fragments serve as seeds for formation of amyloid fibrils elsewhere, and previous work by us suggests that amyloid disaggregation is actually more important than amyloid fibril growth."
Also, knowing how amyloid fibrils are broken up could help researchers to develop treatments for prion and neurodegenerative diseases.
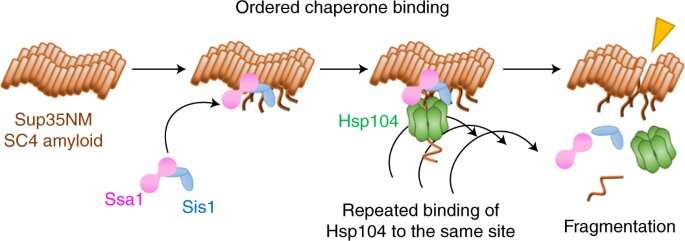
Now, Yoshiko Nakagawa, Tanaka and co-workers have used a form of single-molecule fluorescence microscopy to film how three molecules coordinate to break up amyloid fibrils in a prion system in yeast.
They found that the break-up begins with two chaperones, Hsp70 and Hsp40, preparing the ground by binding to the fibril. A third chaperone, Hsp104, then chips away at the fibril by repeatedly binding to the same site.
Interestingly, the team found that in addition to this fragmentation mechanism, a dissolution mechanism also breaks up fibrils but by a very different chaperone choreography. Which mechanism occurs depends on the molecular shape, or confirmation, of the fibril.
There were many surprises in this study, Tanaka notes. "We didn't expect that Hsp104 attaches to the same site so many times to fragment amyloid fibrils," he says. "And the disaggregation modes for the two different fibril confirmations are very different: the fragmentation mechanism produced many tiny fragments, whereas the amyloid fiber fluorescence gradually disappeared for the dissolution mechanism."
The team intends to explore the mechanisms in greater detail, and in the future they hope to achieve atomic resolution.
More information: Yoshiko Nakagawa et al, Amyloid conformation-dependent disaggregation in a reconstituted yeast prion system, Nature Chemical Biology (2022). DOI: 10.1038/s41589-021-00951-y
Journal information: Nature Chemical Biology
Provided by RIKEN