New insights into the structures and mechanisms of key proteins involved in microbial photosynthesis
In a study published in Nature Communications, scientists from Institute of Biophysics of the Chinese Academy of Sciences, University of Liverpool, Riken Center for Biosystems Dynamics Research, and University of Tokyo, provide new insight into the atomic structures and synthesizing mechanisms of key photosynthetic proteins involved in microbial photosynthesis.
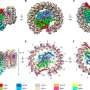
Purple bacteria are the oldest microorganisms on Earth that can harness the energy of sunlight through photosynthesis. The central photosynthetic component in purple bacteria is known as a RC-LH1 core supercomplex, which is formed by a light-harvesting 1 ring (LH1) surrounding the reaction center (RC), together with numerous associated pigment molecules such as bacterial chlorophylls and carotenoids.
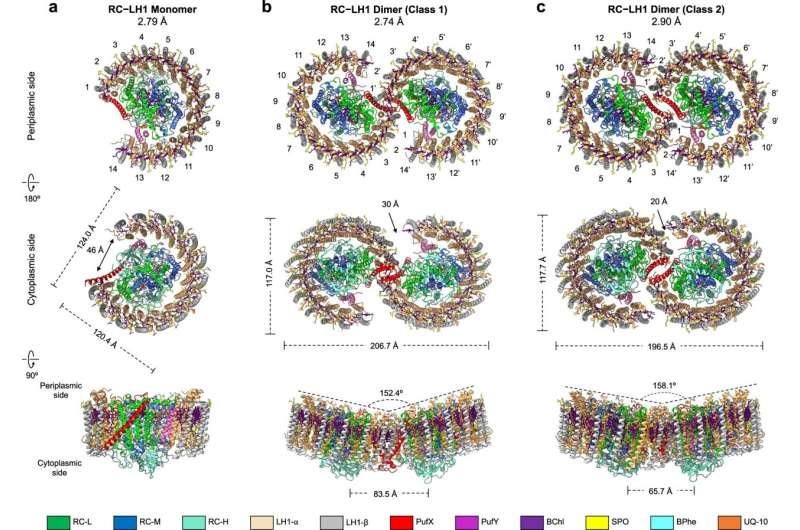
Using cryo-electron microscopy (cryo-EM) and genetic techniques, scientists have uncovered key information about the photosynthetic core complex structure of the purple bacterium Rhodobacter sphaeroides (R. sphaeroides).
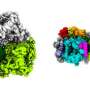
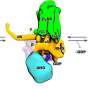
They revealed a dimeric RC-LH1 structure and several structural variants in R. sphaeroides. This RC-LH1 core dimer has an "S-shaped" LH1 ring with a large opening, which is formed by two extra protein peptides known as PufX and PufY. By genetically "deleting" these peptides, they deciphered that PufX is responsible for binding two monomeric cores together to form the dimer structure, whereas the PufY peptide sites at the opposite side of the opening is important to keep the "gate."
Based on the systematic study of the dimeric structure and its variants, scientists proposed how the photosynthetic core complex is generated and self-organized in nature.
They further used computational approaches to prove that the "gates" in the "S-shaped" LH1 ring provide the channels for the electron carrier molecules called quinones to diffuse across the LH1 barrier, which is fundamental for efficient photosynthetic electron transport.
This study provides structural basis for understanding the structural variability and modularity of bacterial photosynthetic complexes, which enable efficient light harvesting, excitation energy transfer, and quinone transport that allow phototrophic bacteria to grow in their specific niche.
More information: Peng Cao et al, Structural basis for the assembly and quinone transport mechanisms of the dimeric photosynthetic RC–LH1 supercomplex, Nature Communications (2022). DOI: 10.1038/s41467-022-29563-3
Journal information: Nature Communications
Provided by Chinese Academy of Sciences