Plant researchers discover cellular mechanism that extends the life of proteins
Plants are tied to one location and need to adjust to their environment, including adverse conditions. Adaptive responses include synthesizing new proteins and breaking down those that are no longer needed. For this task, plants use a considerable amount of energy. Thus, regulation of protein turnover in the plant cell has to be appropriately thorough. Researchers at the Centre for Organismal Studies of Heidelberg University led by Dr. Markus Wirtz and Prof. Dr. Rüdiger Hell have now identified a cellular mechanism that stabilizes proteins by preventing their breakdown.
Plants contain numerous proteins that are needed to adapt to environmental conditions like drought stress. Because their production takes a lot of energy—about half of their energy requirement—plants must precisely regulate protein turnover. To understand how this occurs, the Heidelberg researchers, in collaboration with partners from the Max Planck Institute of Biochemistry in Martinsried and Sapienza University in Rome (Italy), studied thale cress (Arabidopsis thaliana). Thale cress belongs to the family of Brassicaceae and serves as a reference plant due to its short lifetime and simple genome.
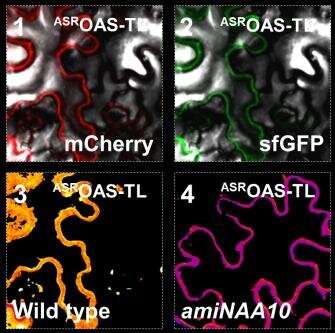
According to the researchers, plants tag proteins while they are synthesized by attaching an acetic acid residue at the beginning of the protein. This process affects more than 80 percent of all plant proteins and is known as N-terminal acetylation. Despite its prevalence, the impact of acetylation was unclear for a long time. The Heidelberg researchers were now able to demonstrate that it protects a majority of the proteins from breaking down through the so-called proteasome, hence prolonging their life. "The proteasome is a molecular shredder that degrades damaged or no longer needed proteins into their building blocks for re-use," explains Prof. Hell.
In genetically altered plants with a reduced protein acetylation rate, Dr. Wirtz's team verified substantially increased proteasome activity, accompanied by accelerated protein degradation. To the astonishment of the researchers, however, the total quantity of proteins within the plant cells remained unchanged. Markus Wirtz: "Plants compensate for the loss of non-acetylated proteins by synthesizing new proteins, probably to dynamically adapt the protein composition in response to environmental stimuli."
The plant researchers' findings may be applicable to human cells. They also have a very similar mechanism to chemically modify and acetylate numerous proteins. "Acetylation appears to be an ancient mechanism that arose several billion years ago in the earliest ancestors of all eukaryotic organisms—organisms which have a true nucleus and a high degree of sub-cellular compartmentalisation," says Prof. Hell.
The German Research Foundation is funding the basic research on protein acetylation in Heidelberg. The results of the study were published in the journal Nature Communications.
More information: Eric Linster et al, Cotranslational N-degron masking by acetylation promotes proteome stability in plants, Nature Communications (2022). DOI: 10.1038/s41467-022-28414-5
Journal information: Nature Communications
Provided by Heidelberg University