Scientists build on AI modelling to understand more about protein-sugar structures
New research building on AI algorithms has enabled scientists to create more complete models of the protein structures in our bodies—paving the way for faster design of therapeutics and vaccines.
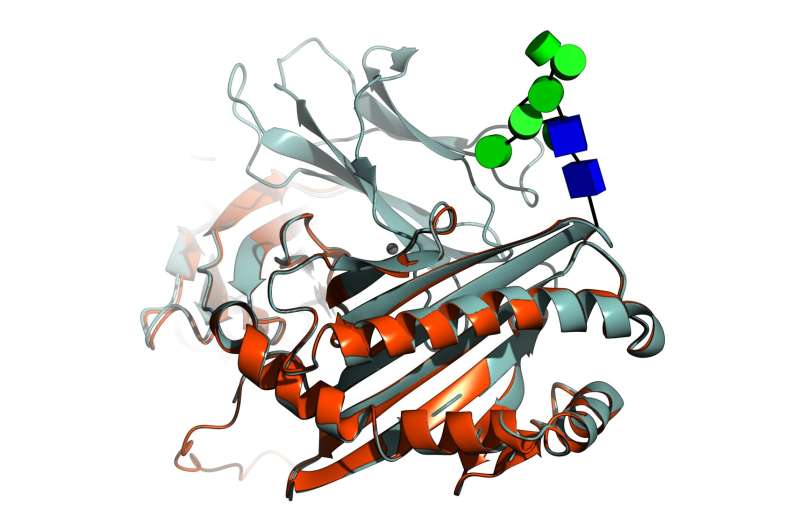
The study—led by the University of York—used artificial intelligence (AI) to help researchers understand more about the sugar that surrounds most proteins in our bodies.
Up to 70 percent of human proteins are surrounded or scaffolded with sugar, which plays an important part in how they look and act. Moreover, some viruses like those behind AIDS, Flu, Ebola and COVID-19 are also shielded behind sugars (glycans). The addition of these sugars is known as modification.
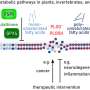
To study the proteins, researchers created software that adds missing sugar components to models created with AlphaFold, which is an artificial intelligence program developed by Google's DeepMind which performs predictions of protein structures.
Senior author, Dr. Jon Agirre from the Department of Chemistry said: "The proteins of the human body are tiny machines that in their billions, make up our flesh and bones, transport our oxygen, allow us to function, and defend us from pathogens. And just like a hammer relies on a metal head to strike pointy objects including nails, proteins have specialized shapes and compositions to get their jobs done."
"The AlphaFold method for protein structure prediction has the potential to revolutionize workflows in biology, allowing scientists to understand a protein and the impact of mutations faster than ever."
"However, the algorithm does not account for essential modifications that affect protein structure and function, which gives us only part of the picture. Our research has shown that this can be addressed in a relatively straightforward manner, leading to a more complete structural prediction."
The recent introduction of AlphaFold and the accompanying database of protein structures has enabled scientists to have accurate structure predictions for all known human proteins.
Dr. Agirre added: "It is always great to watch an international collaboration grow to bear fruit, but this is just the beginning for us. Our software was used in the glycan structural work that underpinned the mRNA vaccines against SARS-CoV-2, but now there is so much more we can do thanks to the AlphaFold technological leap. It is still early stages, but the objective is to move on from reacting to changes in a glycan shield to anticipating them."
The research was conducted with Dr. Elisa Fadda and Carl A. Fogarty from Maynooth University. Haroldas Bagdonas, Ph.D. student at the York Structural Biology Laboratory, which is part of the Department of Chemistry, also worked on the study with Dr. Agirre.
The paper is published in Nature Structural & Molecular Biology.
More information: Bagdonas, H. et al, The case for post-predictional modifications in the AlphaFold Protein Structure Database, Nat Struct Mol Biol (2021). doi.org/10.1038/s41594-021-00680-9
Journal information: Nature Structural & Molecular Biology
Provided by University of York