Accelerating geometry optimization in molecular simulation
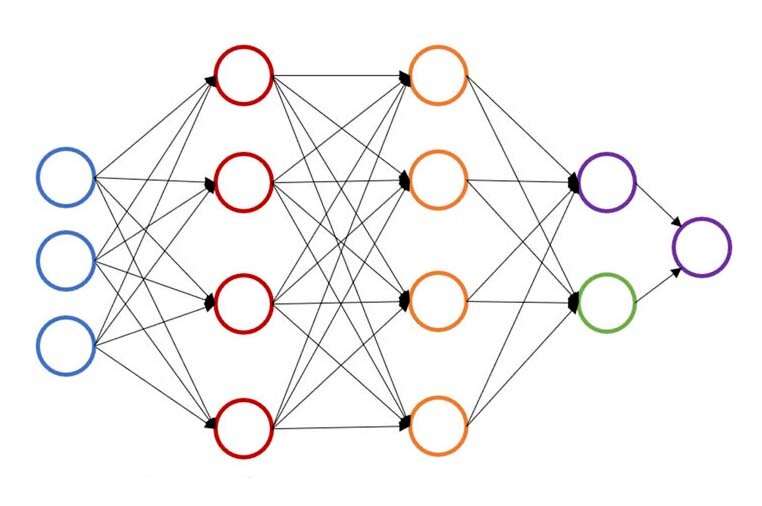
Machine learning, a data analysis method used to automate analytical model building, has reshaped the way scientists and engineers conduct research. A branch of artificial intelligence (AI) and computer science, the method relies on a large number of algorithms and broad datasets to identify patterns and make important research decisions.
Applications of machine learning techniques are emerging in the area of surface catalysis, enabling more extensive simulations of nanoparticles, studies of segregation, structure optimization, on-the-fly learning of force fields, and high throughput screening. However, working through large amounts of data can often be a long and computationally expensive task.
Geometry optimization, often the rate-limiting step in molecular simulations, is a key part of computational materials and surface science. It allows researchers to find ground state atomic structures and reaction pathways, properties used to estimate the kinetic and thermodynamic properties of molecular and crystal structures. While vital, the process can be relatively slow, requiring a large number of computations to complete.
At Carnegie Mellon University, John Kitchin is working to speed up that process by providing a neural network-based active learning method that accelerates geometric optimization for multiple configurations simultaneously. The new model lowers the number of density functional theory (DFT) or effective medium theory (EMT) calculations by 50 to 90 percent, allowing researchers to do the same work in less time or more work in the same amount of time.
"Normally, when we do geometry optimization, we start from scratch," said Kitchin. "The calculations rarely benefit from anything we knew in the past."
"By adding a surrogate model to the process, we have enabled it to rely on previous computations, rather than starting from scratch each time."
The study illustrates the acceleration on several case studies, including surfaces with adsorbates, bare metal surfaces, and nudged elastic band for two reactions. In each case, the Atomic Simulation Environment (ASE)-optimizer Python package allowed for fewer DFT calculations than the standard method.
The ASE-optimizer Python package has been made available to fellow engineers and scientists to make the usage of neural network ensemble active learning for geometry optimization easier.
More information: Yilin Yang et al, Machine-learning accelerated geometry optimization in molecular simulation, The Journal of Chemical Physics (2021). DOI: 10.1063/5.0049665
Github link for the package: github.com/lmj1029123/SingleNN
Journal information: Journal of Chemical Physics