Researchers develop a sequence analysis pipeline for virus discovery
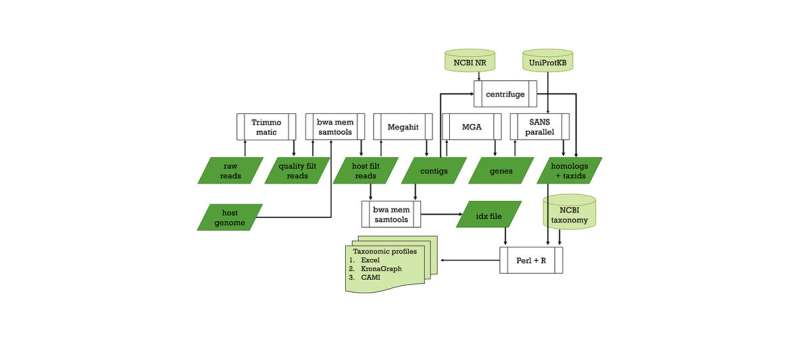
Researchers from the University of Helsinki have developed a novel bioinformatics pipeline called Lazypipe for identifying viruses in host-associated or environmental samples.
The pipeline was developed in close collaboration between virologists and bioinformaticians. The researchers believe they have succeeded to address many challenges typically encountered in viral metagenomics.
Previously,the Viral Zoonooses Research Unit, led by Professor Olli Vapalahti, has published two examples of novel and potentially zoonotic viral agents that were identified with Lazypipe from wild animals that can serve as vectors. A new ebola virus was identified from feces and organ samples of Mops condylurus bats in Kenya, and a new tick-borne pathogen, Alongshan virus, from ticks in Northeast Europe.
"These examples demonstrate the efficacy of Lazypipe data analysis for NGS libraries with very different DNA/RNA backgrounds, ranging from mammalian tissues to pooled and crushed arthropods," says Dr. Teemu Smura.
COVID-19 heightens the need to detect new viruses rapidly
The current Coronavirus pandemic heightens the need to rapidly detect previously unknown viruses in an unbiased way.
"The detection of SARS-CoV-2 without reference genome demonstrates the utilityof Lazypipe for scenarios in which novel zoonotic viral agents emerge and can be quickly detected by NGS sequencing from clinical samples," says Dr. Ravi Kant.
In early April, the research group tested libraries of SARS-CoV-2 positive samples with Lazypipe.
"We confirmed that thepipeline detected SARS-CoV-2 in 9 out of 10 libraries with default settings and without the SARS-CoV-2 reference genome," says Dr. Ilja Pljusnin.
"Lazypipe could play a crucial role in prediction of emerging infectious diseases," adds Assoc. Prof. Tarja Sironen.
More information: Ilya Plyusnin et al. Novel NGS Pipeline for Virus Discovery from a Wide Spectrum of Hosts and Sample Types, Virus Evolution (2020). DOI: 10.1093/ve/veaa091
Provided by University of Helsinki