Precision genome editing enters the modern era
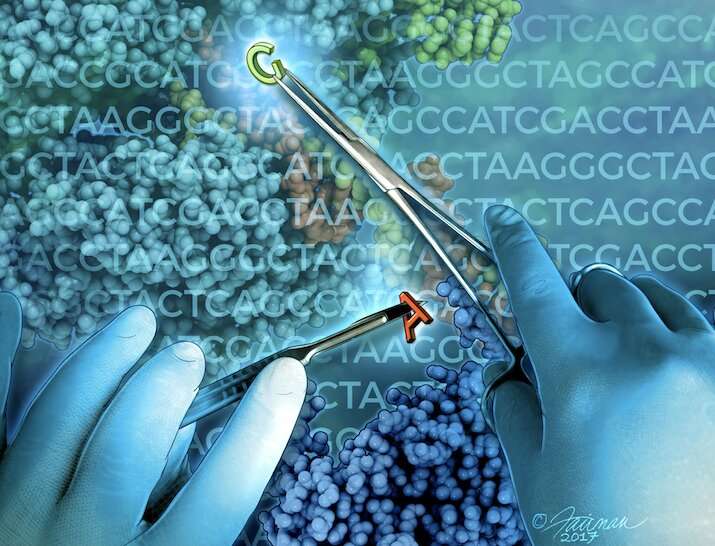
CRISPR has sparked a renaissance in genome editing. Now, next-generation CRISPR technologies let scientists modify the genome more efficiently and precisely than before. Such tools could one day serve as therapeutics, but many challenges remain.
During an unusually warm week in January, a hundred scientists from around the world converged in Palm Springs, California, to talk shop. The biologists, geneticists, and chemists came from start-ups and universities, and they had a lot to discuss. Their topic was the genome—or, more precisely, how to alter it.
It was the 1st International Conference on Base Editing, and the mood in the room was electric, says David Liu, a Howard Hughes Medical Institute (HHMI) Investigator at Harvard University. "The meeting was bustling, people were excited, and it was packed," he says.
In between talks, scientists ducked outside to eat lunch in the sunshine and share the latest in a field recently kindled and currently blazing. Just three and a half years earlier, Liu and colleagues published a paper describing the first "base editor," a tool with the promise of taking the genome editing technology CRISPR to the next level.
With base editors, scientists suddenly had a more efficient and precise way to modify the genome. Such editors could let scientists correct single-letter mutations in DNA—the kind of tiny genetic spelling errors that underlie thousands of human diseases, including sickle cell anemia.
"That was the breakthrough paper," says Reuben Harris, an HHMI Investigator at the University of Minnesota, who helped organize the January conference. "It triggered a landslide in this area." Today, scientists have published more than 300 papers on the technique, used in organisms ranging from bacteria to goats.
Now, the field is inching closer to the clinic. Within the last few months, Harris, Liu, and others have reported the kind of practical refinements needed before base editors are ready for therapeutic use.
Already, scientists have seen success in animals. A week and a half after the conference in Palm Springs, Liu jetted to another meeting, in Banff, Canada, called "Engineering the Genome." This time, he reported using base editors in mice to correct the genetic error behind progeria—a rare human condition characterized by rapid aging and early death. It's a long-term study that Liu's team and their collaborators at the National Institutes of Health and Vanderbilt University are still wrapping up. But he calls the results so far "incredibly exciting."
And this month, he and colleagues report performing a similar feat in mice with a genetic form of deafness. By injecting base editors into the animals' inner ears, the researchers partially restored hearing, Liu's team reports June 3, 2020, in the journal Science Translational Medicine.
Still, many hurdles remain if genome editors are to be used in people—including how to ensure precision, so they revise only where intended, and how to safely deliver them as treatments. Liu acknowledges the field's growth and its challenges. "We are at the fragile beginnings of this new era of human genome editing," he says.
How do base editors alter the genome?
To understand how base editors work, it helps to start with CRISPR.
The powerful genome editing technology burst onto the scene in 2012, offering scientists a new—and simpler—way to make targeted changes in DNA. Until then, scientists relied on clunkier tools. Engineered proteins, for example, like zinc-finger nucleases and TALENs, can edit the genome, but designing them can be costly and time-consuming.
With CRISPR, scientists don't need to create a whole new protein to edit a DNA target. Instead, they can simply generate a short strand of RNA. This "guide" RNA directs its partner, protein scissors called Cas9, to the desired spot. Then, Cas9 tethers itself to the DNA and snips both strands. It's a very efficient way to disrupt a gene, Liu says, because in most cells, cutting DNA leads to chunks of DNA being inserted or deleted at the target site.
CRISPR's potential captured his team's interest. Liu and his new postdoc at the time, Alexis Komor, thought they could harness the technology to do even more—to individually edit the four letters (A, T, G, and C) that make up DNA. "What fascinated me was whether we could make precise edits by doing chemistry on the genome—without cutting the DNA."
Komor and Liu's 2016 breakthrough was linking a version of Cas9 with an enzyme able to rearrange atoms. The result was a hybrid protein, dubbed a cytosine base editor, that could park at specific points in the genome and convert one type of DNA letter into another—C to T or G to A. One year later, Liu's team developed adenine base editors, which can switch A to G and T to C.
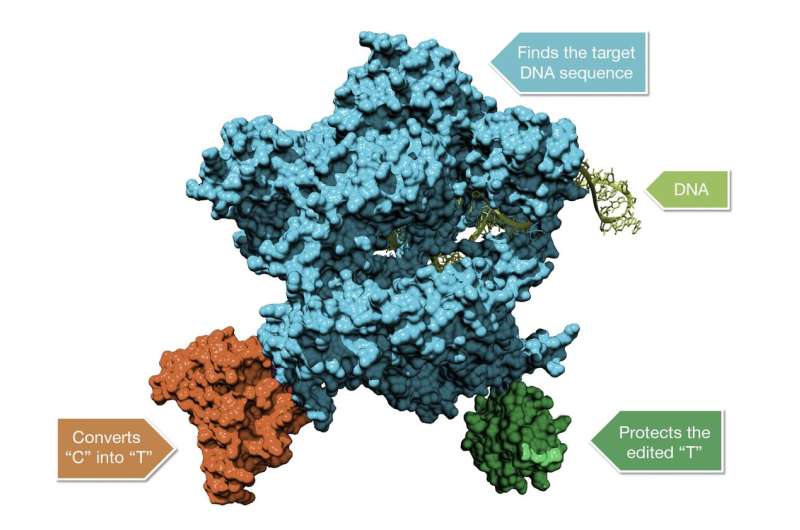
Both classes of base editors fuse two powerful proteins to create an entirely new way to edit the genome, Harris says. "Putting [the base-modifying enzyme] together with Cas9 is sort of like whoever figured out how to put peanut butter together with chocolate," he says. "You come up with something that's even more unique than either alone."
DNA has four "letters." Can base editors swap any one letter for another?
Most base editors can make only certain kinds of swaps. But an even newer tool can do more. About seven months ago Liu's team introduced "prime editors," another technology to precisely edit the genome in human cells. Prime editors still rely on the DNA-binding ability of Cas9, but this time Liu and his postdoc Andrew Anzalone melded it with a different kind of enzyme—one that can insert text directly into the genome.
If Cas9 is like molecular scissors, and base editors are pencils, then prime editors are molecular word processors, Liu says. They do a "search and replace," locating the stretch of DNA researchers want to target and trading the old letters for a stretch of new ones specified by the researcher.
His team has used prime editors to make every possible kind of DNA-letter swap in mammalian cells, in addition to inserting or deleting genomic text. Now, Liu says, "A good chunk of our lab is devoted to advancing prime and base editors to be maximally useful and maximally therapeutically relevant."
How do scientists envision using such precise genome editors?
If you ask Liu about the therapeutic potential of precision genome editors, he'll start giving you numbers.
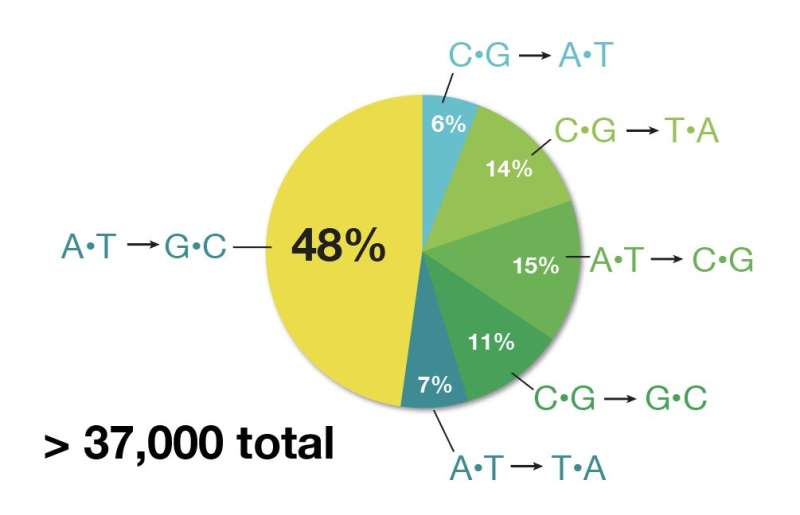
There are more than 75,000 genetic mutations linked to human disease, he says. About half are single-letter DNA swaps. Two-thirds of those swaps are C to T, G to A, or the reverse. Those are exactly the kinds of genetic changes base editors can fix, Liu points out. Such genetic changes, called point mutations, underlie progeria as well as various metabolic disorders, certain causes of deafness and blindness, and hundreds of other genetic conditions. Prime editors offer even more flexibility because they can repair larger genetic glitches. The most common cause of cystic fibrosis, for example, is a small deletion of just three DNA letters.

It all adds up to two new technologies with promise for treating disease. In fact, along with nucleases like Cas9, Liu points out, base editors and prime editors represent the only classes of genome editors known to work in mammalian cells. Neither has made it to clinical trials yet, but with base editors, scientists have had time for some fine-tuning. Work in animals from scientists in academia and industry, Liu says, "has really helped pave the way for base editors' use in patients."
Currently, both base and prime editors are finding use in the lab—to modify genes in cultured cells, for example. And researchers working with plants could potentially use the tools to engineer certain traits, such as resistance to herbicides.
"Both base editing and prime editing are extremely important, especially for agriculture," says Caixia Gao, a plant geneticist at the Chinese Academy of Sciences's Institute of Genetics and Developmental Biology. Her group and Liu's team have now adapted prime editors for use in plants, the researchers reported on March 16, 2020, in the journal Nature Biotechnology.
What challenges remain before base editors are ready for therapeutic use?
In less than four years, base editors have leapfrogged from academic idea to potential therapeutic. But it could be many years—if ever—before such a genetic medicine is ready for market, a fact that Beam Therapeutics, a biotech company Liu cofounded in 2018, pointed out before it went public in February. (The company recently announced preclinical data from its efforts to use base editors to treat sickle cell disease and alpha-1 antitrypsin liver and lung diseases.)
Last year, two groups of researchers from China waved a warning flag. In a pair of papers published in Science, the teams reported that in mouse embryos and rice, cytosine base editors were a little too active; they wielded their red pen more liberally than scientists intended.
Rather than sticking to DNA targets, these base editors sometimes rewrote the genome in unexpected places, says Gao, who led the work in rice. For human use, genome editing has to be just right—"off-target editing" could potentially introduce harmful mutations. (Both papers gave adenine base editors a clean bill of health.)
"We were kind of shocked because we didn't expect to see such effects," Gao says. "We thought, 'here's this extremely important tool, and now we have to reconsider using it.'" Liu sent her an email right away, she remembers. "He said, 'this is a great paper.'"
His team had anticipated the possibility of base editors gone rogue. Liu says the Science papers helped motivate him and others to go after a solution. "I think everyone in the base-editing community felt a responsibility to study and minimize the occurrence of off-target editing," he says.
In February, the researchers published a solution in Nature Biotechnology. They developed a rapid assay to detect the editing errors and created a new suite of cytosine base editors that make 10 to 100 times fewer mistakes. "We wanted to fix the problem," Liu says. The paper was one of a torrent scientists have recently published in the field. Other research groups have independently reported new base editors with very low or undetectable off-target activity. And Liu's team and others have continued to refine base editor technology.
Later that same month, for instance, Harris's team reported in the journal Life Science Alliance another way to achieve greater specificity with cytosine base editors. And Liu's group developed three new Cas9 variants that can target areas of the genome once inaccessible to editing. In June, two other research groups reported dual base editors that combine the DNA letter-swapping abilities of both adenine and cytosine base editors. And Liu's team has now tapped artificial intelligence to predict base editing outcomes, he and his colleagues report June 12, 2020, in the journal Cell.
Base editors still aren't perfect, Harris says. But he calls the spate of recent improvements "several steps in the right direction."
How could scientists eventually package base editors into a form people can use?
Most drugs are small molecules that can be packaged into a pill. Genome editors are large, complicated molecules—so scientists can't just stuff them into a pill for people to swallow, or inject them into people's bodies, Liu says. They have to find other ways to get the molecules into patients' cells.
One method relies on viruses, says Guangping Gao, a gene therapy researcher at the University of Massachusetts Medical School and president of the American Society of Gene and Cell Therapy. Scientists could potentially package genome editors into small viruses like adeno-associated viruses, for example. These viruses, which have already seen clinical use in several FDA-approved drugs, could then infect patients' cells and dump their payloads.
It could be that scientists will need to develop entirely different delivery systems. Researchers are currently experimenting with lipid nanoparticles and using electric fields to coax genome editors into cells that can then be transplanted into patients.
Delivery remains a major hurdle, Gao says, but he's still excited about genome editors' potential. "Gene therapy is now in its golden age," he says. And genome editors "open even more avenues for treating disease."
More information: Mandana Arbab et al. Determinants of Base Editing Outcomes from Target Library Analysis and Machine Learning, Cell (2020). DOI: 10.1016/j.cell.2020.05.037
Wei-Hsi Yeh et al. In vivo base editing restores sensory transduction and transiently improves auditory function in a mouse model of recessive deafness, Science Translational Medicine (2020). DOI: 10.1126/scitranslmed.aay9101
Qiupeng Lin et al. Prime genome editing in rice and wheat, Nature Biotechnology (2020). DOI: 10.1038/s41587-020-0455-x
Jennifer L McCann et al. MagnEdit—interacting factors that recruit DNA-editing enzymes to single base targets, Life Science Alliance (2020). DOI: 10.26508/lsa.201900606
Shannon M. Miller et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs, Nature Biotechnology (2020). DOI: 10.1038/s41587-020-0412-8
Jordan L. Doman et al. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors, Nature Biotechnology (2020). DOI: 10.1038/s41587-020-0414-6
Journal information: Science Translational Medicine , Nature Biotechnology , Science , Cell
Provided by Howard Hughes Medical Institute