March 5, 2019 feature
Nano-bio-computing lipid nanotablet
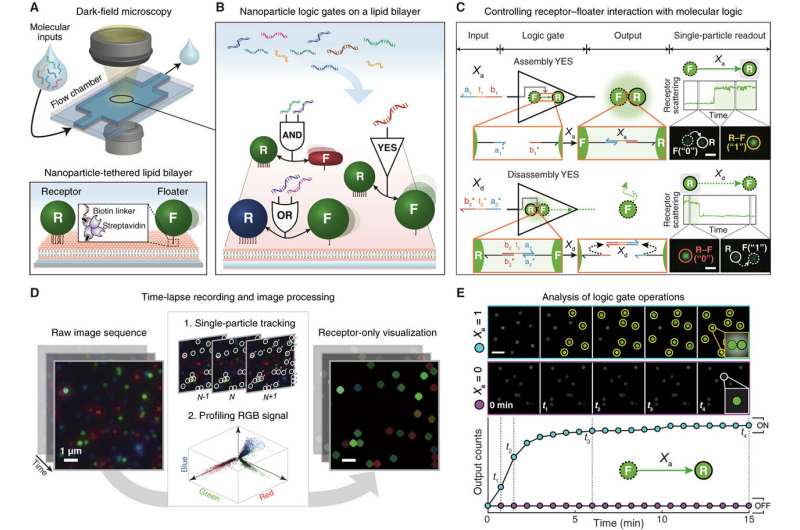
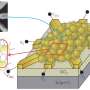
Nanoparticles can be used as substrates for computation, with algorithmic and autonomous control of their unique properties. However, scalable architecture to form nanoparticle-based computing systems is lacking at present. In a recent study published in Science Advances, Jinyoung Seo and co-workers in the Department of Chemistry at Seoul National University in South Korea, reported on a nanoparticle platform built in with logic gates and circuits at the level of the single particle. They implemented the platform on a supporting lipid bilayer. Inspired by cellular membranes in biology that compartmentalize and control signaling networks, the scientists called the platform "lipid nanotablet" (LNT). To conduct nano-bio-computing, they used a lipid bilayer as a chemical circuit board and the nanoparticles as units of computation.
On a lipid nanotablet in solution, Seo et al. established that a single nanoparticle logic gate senses molecules as inputs and triggered particle assembly or disassembly as an output. They demonstrated Boolean logic operations alongside fan-in/fan-out of logic gates and a combinational logic circuit as a multiplexer in the study. The scientists envision the novel approach would be able to modulate nanoparticle circuits on lipid bilayers to engineer new paradigms and gateways in molecular computing, nanoparticle circuits and system nanoscience, in the future.
Matter can be merged with computation across many length scales, ranging from micro-sized droplets in microfluidic bubble logic and micro-particles to biomolecules and molecular machines. Implementing computation in nanoparticles remains unexplored, despite a wide range of applications that could benefit from the ability to algorithmically control the useful photonic, electrical, magnetic, catalytic and material properties of nanoparticles. These properties are currently inaccessible via molecular systems. Ideally, systems of nanoparticles equipped with computing capabilities can form nanoparticle circuits to autonomously perform complex tasks in response to external stimuli to combine the flow of matter and information at the nanoscale.
An existing approach to use nanoparticles as substrates for computation is to functionalize the particles with stimuli-responsive ligands. A group of such modified nanoparticles will then perform elementary logic operations that respond to a variety of chemical and physical inputs. Scientists aim to use an individual nanoparticle as modular nano-parts and implement a desired computation in a plug-and-play manner. However, there are difficulties in wiring integrated multiple logic gates in the solution phase since it is challenging to control the diffusion of inputs, logic gates and output in 3D space. To solve this challenge, scientists were inspired by the cell membrane; a biological equivalent of a circuit board that can host a variety of receptor proteins as computational units. In nature, compartmentalized proteins interact with receptors as a network to conduct complex functions. The membranes can also allow parallel computing processes to occur and therefore materials scientists were inspired to rewire the biological phenomenon.

Bioinspired by cellular membranes, in the present study, Seo et al. demonstrated a lipid bilayer-based nanoparticle computing platform. As a proof-of-principle, they used light-scattering plasmonic nanoparticles to build circuit components, DNA as surface ligands and molecular inputs alongside biotin-streptavidin interactions to tether the nanoparticles to the lipid bilayer. After fixing the nanoparticles to a supported lipid bilayer (SLB), they provided several key features in the experiments;
- They compartmentalized the nanoparticles from a solution containing molecular inputs.
- Particle-to-particle interactions were confined so they would occur only through lateral diffusion at the fluidic 2D reaction space,
- They tracked the laterally confined nanoparticles and analyzed them in situ with single-particle resolution since a large number of light-scattering nanoparticles were shown to be confined in the focal plane using dark-field microscopy (DFM).
The scientists implemented nano-bio computation at the interface of nanostructures and biomolecules, where the molecular information in solution (input) was translated into a dynamic assembly/disassembly of nanoparticles on a lipid bilayer (output). As a key component of an LNT, Seo et al. engineered a flow chamber with a lipid bilayer coated at the bottom of the substrate.

To construct lipid nanotablets in the experimental setup, the scientists used three key components – small unilamellar vesicles (SUVs), glass flow chambers and DNA-functionalized plasmonic nanoparticles. The DNA modified nanoparticles adhered to the lipid bilayer to form logic gates and circuits that processed molecular information. The scientists classified the functionalized nanoparticles into immobile receptors (reporters for computation) or mobile floaters (information carriers of computation). In this context, floaters were "wires" that carried information of upstream gates into downstream gates through robust lateral diffusion. They characterized the nanoparticles to validate their material properties prior to constructing the experimental circuits.
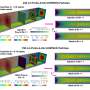
Seo et al. used dark field microscopy (DFM) imaging to measure the performance of the nanoparticle logic gates in response to molecular inputs in solution. When dark-field image sequences were obtained from the logic operations, the scientist processed and quantified them using a custom-built image analysis pipeline.
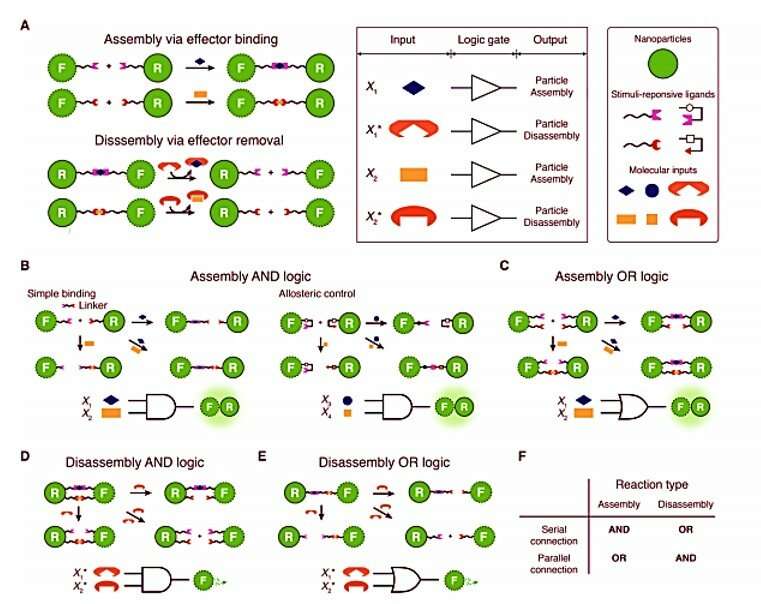
Altogether, the scientists engineered nanoparticle Boolean logic gates and single-nanoparticle YES gates assembly and disassembly operations in real-time. Single-nanoparticle YES gates formed the simplest examples in the study. To detect the scattering signals of a nanoparticle logic gate, the scientists relied on plasmonic coupling between two core particles that composed the gate. To form the nanoparticles, Seo et al. synthesized gold nanorods with silver shells, gold nanospheres and silver nanospheres on gold seeds referred to as red, green and blue nanoparticles to exhibit red, green and blue scattering signals in the study. The scientists represented the behavior of logic-gated nanoparticles in a simple, nanoparticle reaction graph to show an assembly reaction from a floater to a receptor and a disassembly reaction, providing an intuitive view on each nanoparticle logic gate behavior.
The scientists used a sufficiently high density of nanoparticles and incorporated single-particle tracking algorithms to profile the scattering signals and visualize the receptor signals alone in a dark background. To qualitatively understand the overall computing performance of a single nanoparticle logic gate, they used the "receptor-only" view. The results showed that the population of nanoparticle logic gates switched into the ON state in response to performing a YES logic operation. The scientists deduced that a population of nanoparticle logic gates produced high output counts only when the molecular input met TRUE conditions.
To demonstrate two-input, single-nanoparticle logic gates, Seo et al. similarly developed: Assembly AND, Assembly OR, Disassembly AND, and Disassembly OR gates via "interface programming". The scientists showed that the design principles for interface programming were straightforward and could be generalized among circuits. They expanded the interface programming to enable nanoparticle logic gates to process INHIBIT logic.
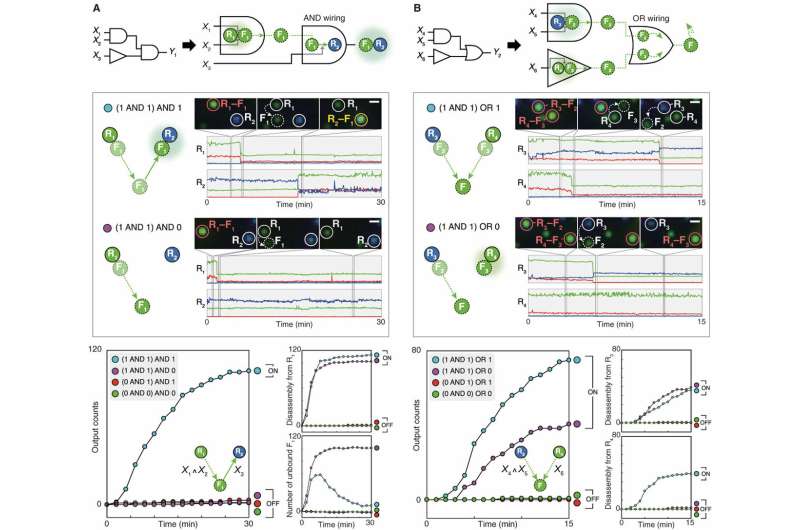
The scientists then increased the complexity of reactions at the receptor-floater interface but noticed incomplete reactions or spurious interactions occuring in the system. Such anomalous interactions indicated that they could not rely on programming particle interfaces as before to construct complex circuits. Instead, they introduced a conceptually distinct approach termed nanoparticle "network programming" to allow two single-particle logic gates to be combined with AND or, OR logic. In the resulting network programming of wired nanoparticle logic gates, the scientists showed the strategy could be implemented to build complex multilayer cascades readily without extensive optimization. Seo et al. successfully implemented the nanoparticle multiplexer to show the ability to design and operate nanoparticle circuits on LNTs in a highly modular and controlled manner.
Scientists can expand on the demonstrated scope of lipid bilayer-based nanoparticle computation to advance the existing molecular computing technologies to operate nanoparticle circuits. They can also integrate lipid bilayers with DNA nanostructures to open the development of new molecular circuits by expanding on dynamic inter-origami interactions for more complex and practical molecular computations. Current limits of the experimental setup prevent the construction of arbitrarily large circuits. These can be overcome to generate broader design space for circuit buildup with new modes of communication, dynamic reconfiguration and DNA walkers.
Seo et al. envision that the molecular computing network can be analogously built in a similar approach to silicon-based computers that have improved through the years. The scientists can advance the experimental setup by increasing the nanoparticle density, to increase the computing capacity and expand parallelism, so that each nanoparticle may independently perform its own computation. For practical applications, the lipid nanotablet will play a pivotal role in building dynamic, autonomous nanosystems in molecular diagnostics and smart sensors; to sense multiple stimuli and trigger the appropriate response. If such nanocircuits are introduced into living cell membranes, scientists can create novel bioengineered nano-bio interfaces as biologic-inorganic hybrid systems. The particles can also be used separately to study membrane-associated phenomena in living cells. In this way, by facilitating communication between nanosystems and cellular systems, the scientists will be able to activate new pathways to navigate complex and dynamic theranostic applications.
More information: Jinyoung Seo et al. Nano-bio-computing lipid nanotablet, Science Advances (2019). DOI: 10.1126/sciadv.aau2124
Maxim P. Nikitin et al. Biocomputing based on particle disassembly, Nature Nanotechnology (2014). DOI: 10.1038/nnano.2014.156
Mikhail Motornov et al. "Chemical Transformers" from Nanoparticle Ensembles Operated with Logic, Nano Letters (2008). DOI: 10.1021/nl802059m
Katarzyna P. Adamala et al. Engineering genetic circuit interactions within and between synthetic minimal cells, Nature Chemistry (2016). DOI: 10.1038/nchem.2644
Journal information: Nature Nanotechnology , Science Advances , Nano Letters , Nature Chemistry
© 2019 Science X Network