Researchers use DNA nanomachines to discover subgroups of lysosomes

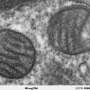
The story of the lysosome is a classic smear campaign. Once dismissed as the garbage disposal of the cell—it does break down unneeded cell debris—it is now valued by scientists who realized all that dirty work also controls survival, metabolism, longevity and even neurodegenerative diseases.
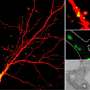
An innovative tool invented by University of Chicago scientists will give us a new window into the lysosome's inner workings. Two studies led by Professor of Chemistry Yamuna Krishnan built tiny machines to tease out clues about lysosomes, including whether they actually come in two or more related types—which may help us understand lysosome-related disorders.
"Both scientists studying the cell and doctors treating patients for lysosome disorders need better diagnostics, so this is a very good step forward," said graduate student Kasturi Chakraborty, the co-first author for both papers.
Scientists want the ability to watch live footage of what's going on in a cell, but its inner workings are hard to catch in action. It's tiny, and what's more, it's a harsh environment; lysosomes in particular are highly acidic—not good for cameras. "Most sensors will just stop functioning if the acidity is that high," Chakraborty said.
To address this issue, Krishnan's group uses DNA as their building material to make flashlights and sensors to peer inside. It's already adapted to life in a cell, and it comes in a handy puzzle-piece format, perfect for building tiny nano-machines that catalogue life inside a living cell.
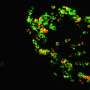
They designed the nanomachines to measure both pH and the particular ions floating around the lysosome—either calcium or chloride—that are the basis for how lysosomes communicate and carry out their tasks. Through them, scientists can see how lysosomes work—and tease out what's going on when they're not working, in certain diseases or hereditary conditions.
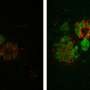
One of the most interesting things they found using the new probes was evidence there are actually at least two different kinds of lysosomes.
Scientists had suspected lysosomes came in distinct types with different functions, but it had never been confirmed, Chakraborty said. They don't yet know exactly how the two kinds of lysosomes differ in function, but they do know one kind is missing in people with a certain lysosome disorder called Niemann-Pick disease.
The key was designing a sensor that could measure two kinds of ions simultaneously. "You absolutely need two independent chemical signatures to discriminate between lysosomes," Chakraborty said.
"It's interesting because lysosomes are well-recognized as a multifunctional organelle, and so til now we considered that it was a single type of lysosome performing multiple functions," said Krishnan, corresponding author for both studies. "Our studies reveal that there might actually be different sub-types of lysosomes designated for different functions."
More information: KaHo Leung et al. A DNA nanomachine chemically resolves lysosomes in live cells, Nature Nanotechnology (2018). DOI: 10.1038/s41565-018-0318-5
Nagarjun Narayanaswamy et al. A pH-correctable, DNA-based fluorescent reporter for organellar calcium, Nature Methods (2018). DOI: 10.1038/s41592-018-0232-7
Journal information: Nature Nanotechnology , Nature Methods
Provided by University of Chicago