Chinese scientists develop novel instrument for rapid profiling of antimicrobial resistance
Widespread antimicrobial resistance (AMR) and the associated rise of "superbugs" is a major public health threat. A leading cause is the misuse or overuse of antibiotics due to the paucity of rapid assays for clinical AMR.
Now, Chinese scientists from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences have introduced a prototype instrument that can directly measure antimicrobial resistant phenotypes at single-cell resolution from clinical samples within three hours, without the need for cell propagation.
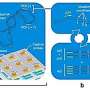
The system, Clinical Antimicrobial Resistance Ramanometry (CAMR-R), is based on a novel approach invented by the team, which showed that AMR can be measured by D2O-feeding Raman Microspectroscopy. When a cell consumes water, in this case D2O or heavy water, the consumption rate can be measured via single-cell Raman spectra. Thus, the level of AMR can be quantified at single-cell resolution, in a culture-free manner.
"All living microbial cells consume water to maintain metabolic activity. Thus, CAMR-R is broadly applicable to different clinical pathogens," said XU Jian, head of the Single-Cell Center of QIBEBT where this system was developed. "CAMR-R provides a solution for the responsible and precise administration of antibiotics."
CAMR-R comes with an intelligent informatics system that includes three pieces of software: CAMR-RamLIS, CAMR-RamEX and CAMR-RamDB, according to XU. CAMR-RamLIS controls rapid, precise and intelligent acquisition of single-cell Raman spectra. CAMR-RamEX performs automated analysis of Raman spectra and computes the distribution of single-cell AMR as well as the average AMR for a clinical sample. CAMR-RamDB supports high-throughput, intelligent identification of pathogens based on the large Clinical Pathogen Reference Ramanome Database.
Traditional AMR tests are based on cell culture and propagation, which measures the degree of microbial inhibition under antibiotics. Such a process typically takes as long as 24 to 48 hours, and is unable to tackle not-yet-cultured or slow-growing pathogens.
"Molecular diagnosis approaches such as nucleic acid detection or genome sequencing can be much faster, but they are applicable only to known gene mutations—not to AMR caused by unknown gene mutations or new mechanisms. Moreover, they are typically unable to quantify the level of AMR," said MA Bo, also from the Single-Cell Center at QIBEBT.
Around the globe, scientists have been racing to develop clinically applicable rapid AMR testing methods. The National Institutes of Health (NIH) of the USA have offered U.S.$20 million in research funding to specifically encourage innovations in this field.
"Employing novel Raman-activated cell sorting technologies invented here, the present CAMR-R system is also capable of sorting those individual microbial cells in the sample that are resistant to antibiotics, so their mechanism of resistance can be revealed via single-cell sequencing. The next generation of CAMR-R will include key clinic-friendly features such as full automation and high-throughput analysis," said XU.
The CAS team has been collaborating with leading hospitals and industrial partners to develop the full clinical diagnosis workflow and facility to target the most important clinical pathogen AMR threats, such as tuberculosis.
"Together, we are laying the foundation for a single-cell clinical AMR monitoring and control network," XU said.
Provided by Chinese Academy of Sciences