Pinpointing mutations that cause bacterial antibiotic resistance
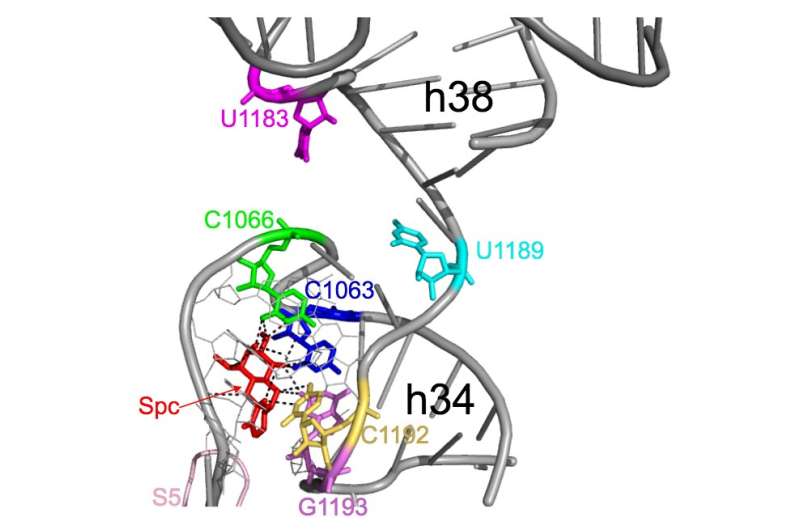
Researchers in Japan have developed a new way of testing bacterial resistance to antibiotics, and found three previously unknown resistance mutations in the process. The fact that they detected both known and unknown mutations suggests their approach will be useful for monitoring resistance to antibiotics.
Antibiotic resistance is a serious problem: more and more pathogenic microorganisms are developing immunity to widely-used antibiotic drugs, rendering them useless. Tremendous effort is being made to identify the mechanisms and mutations that lead to bacterial resistance to antibiotics. Understanding this can help ensure we can effectively treat bacterial infections ranging from pneumonia and infected wounds, to less serious, but contagious conditions such as impetigo and chlamydia.
Researchers from Hokkaido University and the National Institute of Advanced Industrial Science and Technology (AIST) in Japan developed an approach to systematically hunt for resistant ribosomal RNA (rRNA) genes. rRNA is the essential/indispensable part of the cell that creates proteins. It is one of the main targets of antibiotics, but mutations to rRNA is now a well-known route to resistance.
The researchers took rRNA from a wide range of bacterial species in the natural, or non-clinical, environment where mutations are taking place all the time. They inserted them into inactive E. coli lacking in rRNA, and found that more than 2000 imported rRNA could compensate for this lack, therefore preventing the E.coli from dying. They then tested whether a common antibiotic, spectinomycin, effectively killed the bacteria, or if the rRNA gave the E. coli resistance. Their screen found three previously unreported mutations in rRNA from the pathogens that resisted the antibiotic, along with other mutations that were already known. Although using inactive E. coli to test for mutations has been proposed before, this screen can analyse rRNA from other pathogens rather than just what is present in the E. coli.
"Our results suggest that there are many unfound and uncharacterized antibiotic resistance point mutations in rRNA genes," says Kei Kitahara, a molecular microbiologist at Hokkaido University. "We hope our method could be used to verify antibiotic resistance mutations, and contribute to the development of a database for the discovery and diagnosis of resistant bacteria."
The same method could be used to investigate clinical samples, such as testing faecal samples for resistant bacteria in the large intestine, as well as and a broader range of antibiotics.
More information: Kentaro Miyazaki et al. Functional metagenomic approach to identify overlooked antibiotic resistance mutations in bacterial rRNA, Scientific Reports (2018). DOI: 10.1038/s41598-018-23474-4 Kentaro Miyazaki et al. Functional metagenomic approach to identify overlooked antibiotic resistance mutations in bacterial rRNA, Scientific Reports (2018). DOI: 10.1038/s41598-018-23474-4
Journal information: Scientific Reports
Provided by Hokkaido University