At last, a simple way to solve the complex mysteries of the microbiome

This much is clear: The tiny bacteria that live on and inside us are tremendously important for our health and well-being, affecting everything from our mood to the risk of autism. But understanding how those multitudes of microbes interact—and how they influence human health—is a gargantuan task, akin to counting the grains of sand on a beach.
Researchers at the University of Virginia and their collaborators, however, have devised a way to understand not just what is happening but why. By combining cutting-edge computer modeling with old-fashioned laboratory legwork, they have developed a crystal ball to predict how microorganisms will interact and the ripple effects those interactions will have. It's only by understanding this that scientists can hope to manipulate the microbiome, as the organisms are collectively known, to cure disease and improve human health.
"Over the last several years, we have come to appreciate that bacteria living in and on us are critical for a lot of healthy functions," said researcher Jason Papin, Ph.D. But he noted there is far more we don't know than we do know. Too often, scientists find themselves groping around in the dark, especially when they try to engineer the microbiome to benefit health. "Most of what we see is very correlative," Papin said. "We see the presence of the bacteria. What has really been lacking are the actual mechanisms."
The new approach developed by Papin and his collaborators will change that.

Finding Hard Facts
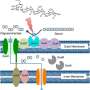
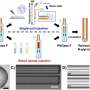
Papin and a Ph.D. student in his lab, Greg Medlock, wanted concrete information: What molecules are being produced by particular bacteria? Which bacteria are then using those molecules? What are the ultimate effects on the populations of different bacteria? The challenge was finding those answers in a system so vast and complex.
The solution was starting small. As Medlock put it, "You kind of have to take a step back."
First Medlock looked at six species of bacteria individually. Then he looked at their interactions in pairs—15 pairs in total. With this data, the researchers were able to develop a computer simulation. The goal was to be able to predict what would happen: As the different species were grown together, would the molecules they produce and consume match up with what was expected? How would these interactions affect the populations of each species?
The researchers then returned to the bench to validate their predictions. The result was hard data that is among the first of its kind, shedding light on the inner workings of the microbiome. But the importance of the research isn't just in the findings but in the approach the scientists have created. They call it an "experimental and computational pipeline," and it opens the door to fast, factual research examining many different aspects of the microbiome. That will have tremendous benefits in the quest to manipulate the microbiome to improve human health. Now, if scientists think increasing a particular type of bacteria would be beneficial, or want to reduce a harmful one, they can determine how they can make that happen. "Once you have this knowledge, you can actually intervene. You can actually control," Papin said. "It becomes at least possible to engineer the system."
More information: Gregory L. Medlock et al, Inferring Metabolic Mechanisms of Interaction within a Defined Gut Microbiota, Cell Systems (2018). DOI: 10.1016/j.cels.2018.08.003
Journal information: Cell Systems
Provided by University of Virginia