Loss of a microRNA molecule boosts rice production

The wild rice consumed by our Neolithic ancestors was very different from the domesticated rice eaten today. Although it is unclear when humans first started farming rice, the oldest paddy fields—in the lower Yangzi River Valley—date back to 4000 BC. During its long history of cultivation, rice plants with traits that reduce yield or impede harvest (e.g., grain shattering) were weeded out, whereas those with traits that increase yield (e.g., highly branched flowering structures) were selected and propagated. Although the resulting rice plants are super-producers that feed much of the world's population, they rely on human assistance and cannot withstand harsh environmental conditions.
Scientists can examine the genetic basis for some of the changes that took place during rice domestication by comparing genes in cultivated rice plants with those in their wild rice relatives. Using this approach, several key genes that were altered during domestication, such as those affecting grain shattering, have been identified and studied. Most of these genes encode transcription factors that bind to other genes and regulate their activity.
A team of researchers from the National Centre for Biological Sciences, Tata Institute of Fundamental Research in India led by Dr. P.V. Shivaprasad wondered whether another type of molecular regulator, named microRNAs, also contributed to the domestication of rice. MicroRNAs regulate specific target genes by binding to RNA copies of the gene and, together with other molecules, blocking their activity or chopping them into tiny fragments. In special cases, the resulting RNA fragments trigger a silencing cascade, shutting down the activity of genes that are similar to the initial target gene.
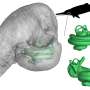
The researchers compared the microRNA populations of high-yielding indica rice lines with those of wild rice and several traditional rice varieties. One microRNA species stood out: miR397 accumulated to high levels in the flag leaves of wild rice, but was barely detectable in the other plants analyzed. The scientists showed that miR397 silenced several members of the laccase gene family via a silencing cascade. Laccase genes, of which there are 30 in the rice genome, encode proteins that promote woody tissue formation, thereby providing mechanical strength. By silencing a subset of these genes, miR397 greatly reduced the formation of woody tissue. Furthermore, when the scientists transgenically expressed the gene encoding miR397 in domesticated rice, the resulting plants were more similar to wild rice plants than to domesticated ones, with long, spindly stems; narrow, short leaves; few flowering structures; and hardly any rice grains. In effect, the team partially de-domesticated rice by increasing the levels of a single microRNA species.
These findings raise intriguing questions. If silencing several laccase genes by increasing miR397 levels negatively affects yield, would upregulating the expression of this same set of laccase genes boost grain production? In addition, would reducing the levels of miR397 in wild rice plants, and thereby lifting the repression of the laccase genes, improve yields, while retaining the traits that allow wild plants to thrive in harsh environments? "miR397 and laccase genes overlap with unknown genomic regions predicted to be involved in rice yield. Modifying their expression in wild species and cultivated rice would be useful in improving yield and other beneficial characters. We hope that our finding promotes future research to identify other changes associated with domestication of plants, spearheading further improvement in crops for the future," states Dr. Shivaprasad.
More information: Chenna Swetha, Debjani Basu, Kannan Pachamuthu, Varsha Tirumalai, Ashwin Nair, Melvin Prasad, and P. V. Shivaprasad. (2018). Major Domestication-Related Phenotypes in Indica Rice are Due to Loss of miRNA-Mediated Laccase Silencing. Plant Cell doi.org/10.1105/tpc.18.00472
Journal information: Plant Cell
Provided by American Society of Plant Biologists