This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Telomere-to-telomere genome assembly research opens the door to new crop varieties
Completely new crop varieties which can better withstand drought, salinity and pests are within reach thanks to international genomics research published today in Nature Genetics.
The study, "Unlocking plant genetics with telomere-to-telomere genome assemblies," was led by researchers at Murdoch University's Center for Crop and Food Innovation (CCFI).
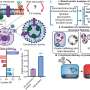
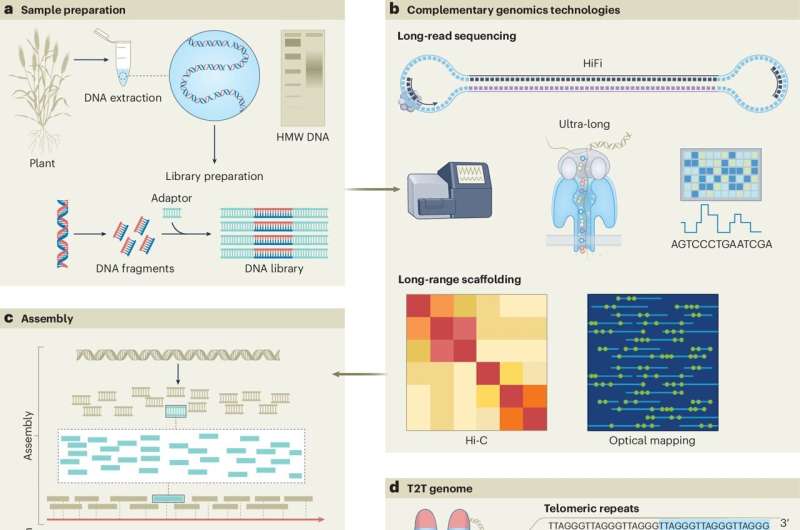
Complete Telomere-to-Telomere (T2T) assemblies allow scientists to map an entire genome to underpin molecular breeding to improve plant performance under stress conditions.
Study lead and CCFI Director, Professor Rajeev Varshney said the genome assemblies' findings represented the future of genetics.
"Genome assembly is essentially like building a jigsaw, piece by piece. T2T genome assemblies provide a complete, end-to-end picture of a genome in its entirety," Professor Varshney said.
"Our job is to make sure that the thousands of genes within a genome are assembled in the correct order, along the entire chromosome, and that each piece fits together perfectly."
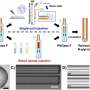
First author and CCFI researcher Dr. Vanika Garg, said the level of accuracy now possible in gene sequencing had led to the breakthrough.
"Until now, T2T genome assemblies have not been possible, and incomplete gene sequencing can lead to errors, such as mis-annotations," Dr. Garg said.
"This means that the genes responsible for certain desirable traits are missed. But thanks to advances in sequencing technologies, we can finally have T2T genome assemblies for any crop species."
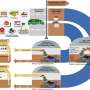
Professor Varshney and his research team are currently working in collaboration with several laboratories from Australia, UK, Germany, U.S., and China on developing T2T genome assemblies or nearly complete genome assemblies for several crops such as wheat, chickpea, fava bean, papaya, passion fruit, custard apple, banana and pineapple.
"Together with our national and international collaborators, we are applying methods of T2T genome assemblies in several crops and are incredibly excited to see how this can advance agricultural research in Australia and abroad," Professor Varshney said.
"T2T genome assemblies grant access to difficult-to-sequence regions, which opens up a myriad of groundbreaking research possibilities, such as creating completely new crop varieties that cater to our future needs."
More information: Vanika Garg et al, Unlocking plant genetics with telomere-to-telomere genome assemblies, Nature Genetics (2024). DOI: 10.1038/s41588-024-01830-7
Journal information: Nature Genetics
Provided by Murdoch University