New computational method for drug discovery
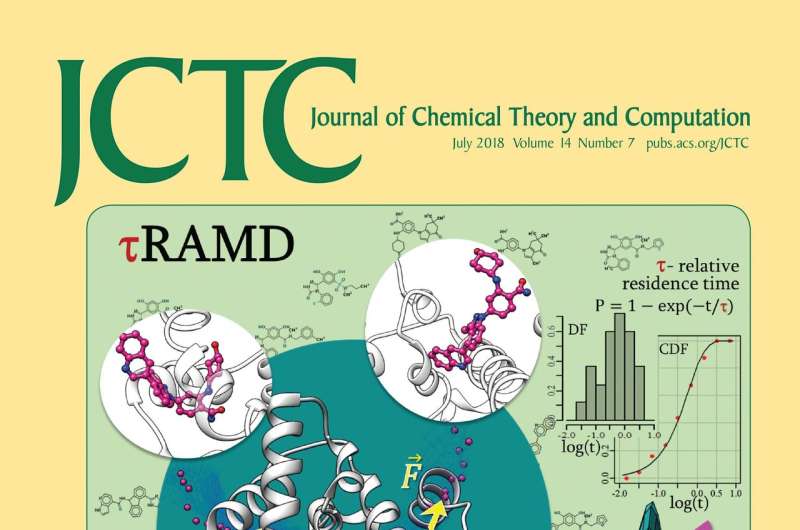
HITS researchers developed tauRAMD, a tool to predict drug-target residence times from short simulations. The method is illustrated on the cover page of July 2018 issue of the Journal of Chemical Theory and Computation, software is freely available.
The design of a drug with a desired duration of action, whether long or short, is usually a complicated and expensive trial-and-error process guided only by a mix of expert intuition and serendipity. One of the parameters affecting drug efficacy is the lifetime of the complex formed between a drug and its target protein, whose function must be altered, e.g. inhibited. In practice, many possible chemical compounds have to be synthesized and then tested to discover an appropriate drug candidate.
Easy method and high performance
As a part of the Kinetics for Drug Discovery (K4DD) project supported by the EU/EFPIA Innovative Medicines Initiative Joint Undertaking, researchers at the Molecular and Cellular Modeling (MCM) group at the Heidelberg Institute for Theoretical Studies (HITS) developed a computationally efficient and easy-to-use method for predicting the relative lifetimes of complexes of a target protein with different drug candidates. The scientists demonstrated the high predictive performance of the computational approach using experimental data measured by collaborators at Merck KGaA (Darmstadt), Sanofi-Aventis Deutschland (Frankfurt am Main), and Sanofi R&D (Vitry-sur-Seine, France).
The method, called tauRAMD (residence time, tau, estimation using Random Acceleration Molecular Dynamics simulations) has been developed for ease of use and makes it possible to compute long residence times with short simulations. It has been successfully applied to diverse sets of compounds binding a range of therapeutically important target proteins. The software is freely available at www.h-its.org/downloads/ramd/.
More information: Daria B. Kokh et al. Estimation of Drug-Target Residence Times by τ-Random Acceleration Molecular Dynamics Simulations, Journal of Chemical Theory and Computation (2018). DOI: 10.1021/acs.jctc.8b00230
Provided by Heidelberg Institute for Theoretical Studies