DIY brings high throughput to continuous cell culturing
Rice University's Caleb Bashor never planned to be an inventor or do-it-yourselfer, but there was no other way to do the microbiology experiments he envisioned.
In a paper appearing online this week and on the cover of July's issue of Nature Biotechnology, Bashor and colleagues from Boston University (BU) and the Massachusetts Institute of Technology unveil eVOLVER, a system that combines the control of automated cell-culturing systems that can run continuously for months with the scale of high-throughput systems that grow dozens of cultures at once.
Bashor, assistant professor of bioengineering at Rice, first thought of the idea for eVOLVER three years ago while working as a postdoctoral researcher at BU.
"One thing we were working on was antibiotic resistance," he recalled. "A lot of times, antibiotic resistance shows up when you administer the drug continuously. So I was thinking, what if you administer it in pulses? How long are the pulses? Where do you see antibiotic resistance evolve? Where do you see it not evolve? Is there a way to systematically see what the temporal features of antibiotic administration gives you?
"If you plan a time course and say, 'I want to do this one every 15 minutes, I want to do this one every hour,' and so on, you quickly realize that it's impossible without automation," he said. "Not to mention that antibiotic resistance often shows up in time scales of weeks rather than days. You need something automated that can run for several weeks, with dozens or hundreds of samples that are individually controlled and monitored."
Bashor, a trained biochemist and synthetic biologist, knew that no off-the-shelf device existed for such an experiment, and combining standard bioreactors in a high-throughput setup would require entire rooms of space and cost hundreds of thousands of dollars.
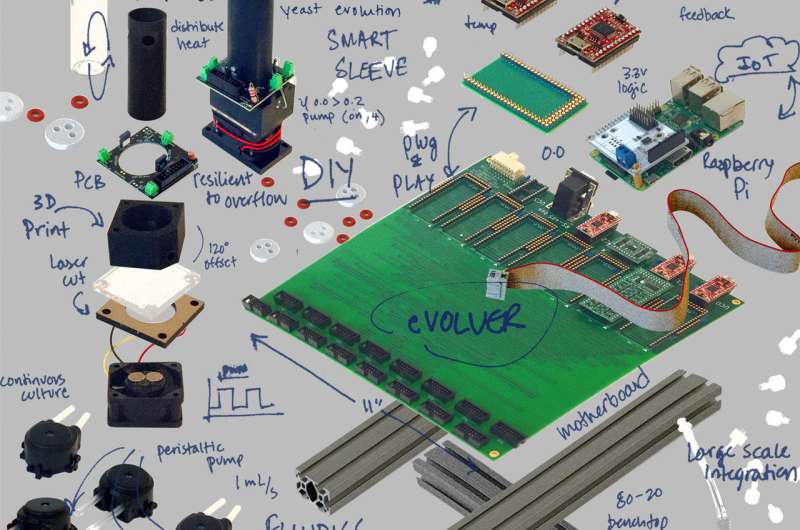
"I was aware of the do-it-yourself electronics culture, and I kept thinking, 'There has to be a way to make this that's not terribly difficult and would be relatively cheap,'" he said. "I started looking at microcontrollers. I started looking online at electronics catalogs and materials, and I thought about, 'Okay, what are the materials we could use?' 'How do we keep this simple?'"
The earliest collaborators on the project were study co-authors Ahmad "Mo" Khalil, assistant professor of bioengineering at BU, and Brandon Wong, a graduate student in Khalil's group. Together, the three and other collaborators worked on eVOLVER in their spare time, designing the electronics, fluidics, culture chambers and the software that would both control each chamber and report results.

"The software was a challenge," Bashor said. "I wrote a little loop in MatLab to control like a half-dozen cultures, and better coders took it from there and elaborated on it. The code got quite enormous actually."
The group struggled to find code that could report, record and track all variables across all time scales in each culture chamber but eventually struck gold with software that was originally designed to track stock prices.
"It allows us to track data, as it accumulates, live, and we can look at the timeline of the data on any resolution. It also stores all the data on a server," he said.
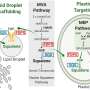
The prototype 16-chamber version of eVOLVER described in the new paper cost less than $2,000, cheaper than what a lab might pay for a single continuous culture bioreactor, Bashor said. eVOLVER comprises three modules: a customizable "smart sleeve" housing and interface for each culture vessel, a fluidic module that controls movement of liquid in and out of each culture vessel, and a modular hardware infrastructure that simplifies high-volume bi-directional data flow by decoupling each parameter into individual microcontrollers.
"The idea was to make it cheap, open source and totally customizable so that anybody could make it as long as they have access to a machine shop, a 3-D printer and a McMaster-Carr catalog or RadioShack catalog," he said.
In proof-of-principle experiments described in the study, the researchers used eVOLVER to individually vary the level and timing of temperature, media inputs and outputs and other features of the environment in more than 450 yeast cultures over a period of 500 hours. Sensors continuously reported how each culture performed, allowing the researchers to gauge how well particular strains of yeast adapted to specific levels of stress.
Bashor said he plans to build an even larger, 96-chamber version of eVOLVER in his own lab at Rice's BioScience Research Collaborative this fall with help from visiting collaborators. While the device won't be used in every experiment he plans for the lab, he is planning for some of the synthetic biology experiments he originally envisioned for eVOLVER.
"We can put synthetic circuitry into the cells, and then we can evolve the cells to see how they change to accommodate the circuitry," he said. "Stress, from factors like heat or starvation, can put pressure on cells to evolve, and putting synthetic circuitry into a cell can act as a pressure too. The cells don't normally have the genes from these circuits, so there's natural pressure to evolve to get rid of them.
"But some of the cells might evolve a way to keep the circuits, and we would want to know how they did that. That would be something we'd want to be able to engineer."
More information: Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER, DOI: 10.1038/nbt.4151 , www.nature.com/articles/Nbt.4151
Journal information: Nature Biotechnology
Provided by Rice University