Researchers film bacteria using melee combat to steal antibiotic resistance genes

Researchers at the University of California San Diego Center for Microbiome Innovation have identified the mechanism by which a clinically relevant bacterium may gain antibiotic resistance, and have come up with a model for predicting the conditions under which it spreads. The findings, which establish a framework for understanding, quantifying and hopefully combating the emergence of superbugs, were published in a recent paper in eLife.
The spread of antibiotic resistance among pathogenic bacteria is a major public health concern. Among the top three most dangerous are Acinetobacter species, which thrive in hospital settings and acquire antibiotic resistance genes surprisingly quickly.
"Most of the threat posed by Acinetobacter stems from its ability to acquire drug resistance via horizontal gene transfer (HGT)," said Jeff Hasty, professor of biology and bioengineering at UC San Diego and principal investigator on the paper.
HGT is the process by which bacteria exchange genetic material. Acinetobacter species do this at a remarkably high rate, which makes them particularly good at becoming resistant to antibiotics.
Until now, HGT has not been directly observed in Acinetobacter species—and it was observed by accident.
When Robert Cooper joined the Hasty lab as a postdoctoral researcher, he was interested in using algae and bacteria to make biofuels.
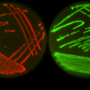
"I was looking for a side project and remembered reading about 'predator' Acinetobacter bacteria that engage in hand-to-hand combat to kill their prey," said Cooper. "They do so by extruding something analogous to a poison-tipped dagger, and stabbing neighboring cells."
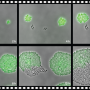
Intrigued, Cooper acquired some "predator" bacteria and placed them next to "prey" E. coli cells. He gave the predators a red and the prey a green fluorescent protein so that he could tell them apart in live-action movies. The "predator" bacteria did kill the E. coli as expected—but they also started glowing green!
The cells in this video are growing inside a microfluidic chip—a microscopic chamber with a channel beneath it to supply fresh nutrients and wash away excess cells. Each oval is a single bacterial cell.
This marked the beginning of a journey for Cooper.
He attached the green fluorescent protein to an antibiotic resistance gene and observed that the when Acinetobacter killed neighboring E. coli cells, those that acquired the green gene also took up and incorporated the antibiotic resistance gene (see video). Those cells were then able to propagate, creating a colony of drug-resistant Acinetobacter.
Cooper went on to establish a model for predicting the number of "prey" cells that would be killed, and how many "predator" cells would acquire the antibiotic resistance gene by HGT, under different conditions.
"The model predicts the efficiency of uptake and incorporation of the antibiotic resistance gene," said Cooper. "We hope that it will be useful for determining the conditions under which antibiotic resistance might spread, which could help us focus on the most important areas for prevention."
More information: Robert M Cooper et al. Inter-species population dynamics enhance microbial horizontal gene transfer and spread of antibiotic resistance, eLife (2017). DOI: 10.7554/eLife.25950
Journal information: eLife
Provided by University of California - San Diego