Enzyme LSD1 found to regulate muscle fiber type differentiation
Our bodies convert food into energy for all life activities. These metabolic processes allow cells to produce energy (anabolism) and consume energy from nutrients (catabolism). In a recent update out of Kumamoto University in Japan to a continuing research project from the Institute of Molecular Embryology and Genetics (IMEG), researchers found that the LSD1 enzyme suppresses the expression of genes involved in mitochondrial metabolism and the slow-muscle genes that cooperate with them.
All cells regulate metabolism-related gene expression and adapt to environmental changes such as nutrition, oxygen supply, exercise, and temperature. Normal cells produce energy using mitochondrial metabolism when oxygen is abundant, and glycolysis (metabolism using mainly sugar) when oxygen is scarce. When metabolism changes between these mechanisms, the function of the cellular metabolic gene groups changes greatly. However, the mechanisms of these changes are still unknown.
Generally, gene function is determined by the state of transcription factors and genome modifications (chemical changes). The modified genome is called an epigenome, and includes DNA methylation and modification of gene-regulating proteins, called histones, which DNA wraps around. Among these modifications, the methylation of histone lysine is an important part of the epigenome. It is created by methylation enzymes attaching methyl groups to histones and demethylating enzymes removing them.
In 2012, the Kumamoto University research group became the first to show the mechanism by which the demethylating enzyme LSD1 regulates energy metabolism in adipocytes (www.nature.com/articles/ncomms1755>Nature Communications). Their research demonstrated the mechanism by which LSD1 suppresses the activity of mitochondrial metabolism genes, resulting in increases fat accumulation and obesity. Furthermore, in 2015, they revealed that LSD1 suppresses mitochondrial metabolism in cancer cells and promotes glycolysis in the presence of oxygen (cancerres.aacrjournals.org/content/75/7/1445.long>Cancer Research). Thus, they showed that LSD1 is an epigenome modification enzyme that regulates metabolism depending on the state of various cells.
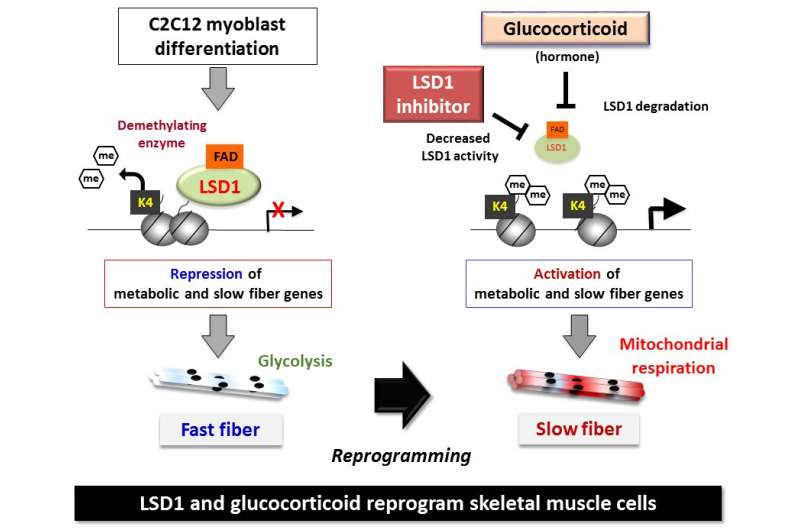
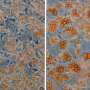
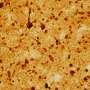
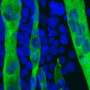
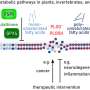
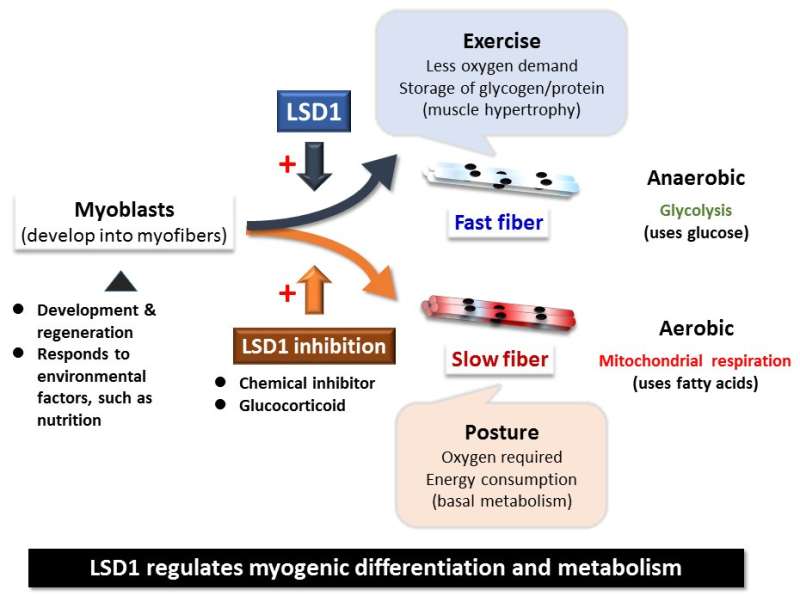
Skeletal muscle acts as a metabolic organ in the body and performs glycolysis in fast muscles and mitochondrial metabolism in slow muscles. In recent work, the researchers used mouse myoblasts (C2C12 cells) to analyze gene expression and the epigenome. They found that LSD1 suppresses both the expression of the genes involved in mitochondrial metabolism and the slow-muscle genes that cooperate with them. LSD1 was found to accumulate in metabolism-related loci and slow-muscle type loci to suppress gene function. Interestingly, the action of glucocorticoid, a major metabolic control hormone, promoted the degradation of LSD1 and reduced the LSD1 protein. In other words, myoblasts differentiate into fast muscle fibers in the presence of LSD1, and slow muscle fibers when LSD1 is suppressed with glucocorticoid. Thus, the relationship between the hormone glucocorticoid and the enzyme LSD1 was shown for the first time.
Researchers also found that a combination of glucocorticoid and LSD1 enhances the function of metabolism-related and slow-muscle type genes. Consequently, LSD1 coordinately controls skeletal muscle differentiation and metabolism under the action of hormones responding to the environment. This is thought to play an important role in skeletal muscle metabolism.

Recently, it has become clear that obesity and diabetes have common metabolic pathologies. Dementia, muscle diseases, and other age-related diseases also result from a functional decline of energy metabolism. In an aging society, the reduction of skeletal muscle mass (sarcopenia) and of mitochondrial metabolism in the skeletal muscle of obese and diabetic patients are drawing attention. The results of this research can be expected to be useful in the development of molecular mechanisms for new control and prevention methods of skeletal muscle dysfunction due to aging.
This research was posted online in the journal Nucleic Acids Research on 29 March 2018.
More information: Kotaro Anan et al, LSD1 mediates metabolic reprogramming by glucocorticoids during myogenic differentiation, Nucleic Acids Research (2018). DOI: 10.1093/nar/gky234
Journal information: Nature Communications , Nucleic Acids Research , Cancer Research
Provided by Kumamoto University