Neutrons observe vitamin B6-dependent enzyme activity useful for drug development
Scientists at the Department of Energy's Oak Ridge National Laboratory have performed neutron structural analysis of a vitamin B6-dependent protein, potentially opening avenues for new antibiotics and drugs to battle diseases such as drug-resistant tuberculosis, malaria and diabetes.
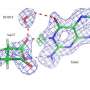
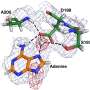
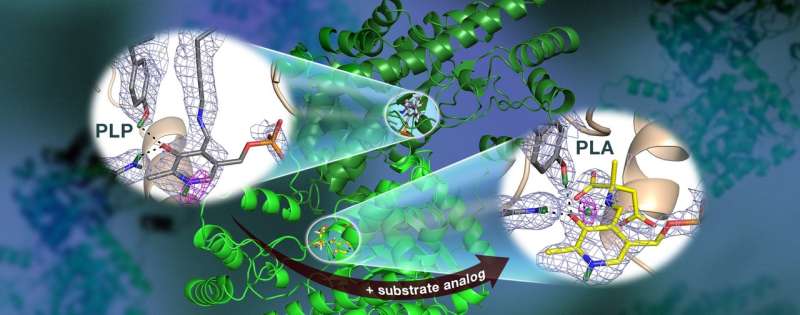
Specifically, the team used neutron crystallography to study the location of hydrogen atoms in aspartate aminotransferase, or AAT, an enzyme vital to the metabolism of certain amino acids.
"We visualized the first neutron structure of a vitamin B6 enzyme that belongs to a large protein family with hundreds of members that exist in nature," said ORNL's Andrey Kovalevsky, a senior co-author of the study, which was published in Nature Communications.
Vitamin B6-dependent proteins are part of a diverse group of enzymes that conduct over a hundred different chemical reactions in cells. The enzymes are of interest to biomedical, as well as bioenergy, researchers because of their role in metabolizing amino acids and other cell nutrients.
"These enzymes are unique in that each one performs a specific chemical reaction with exquisite accuracy, while suppressing other viable chemical transformations," Kovalevsky said. "How they accomplish this is not well understood, but it is of great significance for drug design."
The team's previous research predicted that hydrogen atoms move in and around the enzyme's active site, where the chemical reaction takes place, indicating that the hydrogen atoms' positioning controls the reaction type. Knowing the precise location of hydrogen atoms can explain why the behavior of these enzymes is so specific, but hydrogen is hard to detect with standard methods such as X-ray crystallography.
To directly determine the positions of hydrogen atoms within AAT, the ORNL-led team turned to neutron diffraction techniques. The researchers exposed delicate protein crystals to neutrons using the IMAGINE beamline at ORNL's High Flux Isotope Reactor and the LADI-III beamline at the Institut Laue-Langevin in Grenoble, France.
Surprisingly, the team observed a reaction within one AAT protein biomolecule while another AAT biomolecule was unchanged, providing a before-and-after perspective of the enzyme-catalyzed chemical reaction.
"The data revealed that in one of the enzyme's biomolecular structures the covalent bonds reorganized after a chemical reaction occurred in the active site and, in another, the reaction had not taken place," Kovalevsky said. "Essentially, we were able to obtain two structures in one crystal, which has never been done before for any protein using neutrons."
With this knowledge, the team will run molecular simulations to determine the hydrogen atoms' specific behavior when interacting with the enzyme. The results could be useful in guiding the future design of novel medicines against multidrug-resistant tuberculosis, malaria, diabetes and antibiotic-resistant bacteria.
"This study highlights how neutrons are an unrivaled probe for identifying the location of hydrogen atoms in biological systems, providing us with an unprecedented level of structural detail for this important enzyme," LADI-III beamline scientist Matthew Blakeley said.
More information: Steven Dajnowicz et al, Direct visualization of critical hydrogen atoms in a pyridoxal 5′-phosphate enzyme, Nature Communications (2017). DOI: 10.1038/s41467-017-01060-y
Journal information: Nature Communications
Provided by Oak Ridge National Laboratory