Encoding smart antibiotics

A method for designing antibiotics based on random binary encoding, developed by a team led by the National Physical Laboratory (NPL), could open up new opportunities in drug discovery.
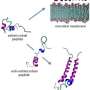
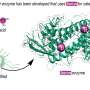
Biological activity is encoded in molecular sequences of twenty unique amino acids. Antimicrobial activity is no exception and is programmed in short sequences called antimicrobial peptides, which are used by our immune systems to combat bacteria.
As the spread of antimicrobial resistance drives the need for stronger, faster and more selective treatments, researchers are developing new sequences based on these naturally-occurring peptides for use in antimicrobial therapies. However, much remains unknown about such sequences – in particular, which sequences are most effective against bacteria without harming our own bodies' cells? And what structural features drive selectivity?
An international team of researchers led by NPL set out to explore antimicrobial selectivity by creating two sets of millions of random antimicrobial sequences built from just two amino acids. The first set of sequences they created was predicted to effectively kill bacteria while also affecting human red blood cells; the second set of sequences was created to exclusively target bacterial cells.
To achieve this, the team capitalised on the property of chirality by replacing one of the two amino acids with its mirror image. All naturally-occurring protein sequences are chiral (i.e. not identical to their mirror image), a property which leads to their reversed-chirality (mirror image) forms not being able to impact on our immune systems. In contrast, bacteria often switch chirality to produce antibiotics capable of fighting off other bacteria, and can therefore be affected by reversed-chirality sequences.
Consequently, the set of antimicrobial sequences with partially-reversed chirality effectively killed bacteria, including superbugs MRSA and VSE, without negatively affecting human cells, even at very high concentrations. Most strikingly, the two sets of sequences exhibited two fundamentally different physical mechanisms – the toxic homochiral sequences tended to perforate bacterial membranes, while the highly-selective reversed-chirality sequences left no visible markings on membrane surfaces.
The findings, reported in the journal Angewandte Chemie and conducted in collaboration with the Hebrew University of Jerusalem, the University of Brighton, the University of Western Australia and the University of Oxford, could open up new opportunities in drug discovery for encoding highly-selective antimicrobials.
Reproducible measurements of antimicrobial activity are essential to ensuring confidence in the next generation of safe, effective treatments, and NPL's Biotechnology group is developing the measurement infrastructure needed to underpin antimicrobial discovery and development.
More information: Zvi Hayouka et al. Binary Encoding of Random Peptide Sequences for Selective and Differential Antimicrobial Mechanisms, Angewandte Chemie International Edition (2017). DOI: 10.1002/anie.201702313
Journal information: Angewandte Chemie , Angewandte Chemie International Edition
Provided by National Physical Laboratory