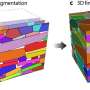
Single cell RNA-seq will provide key insight on how different types of taste cells function

A significant technological advance from the Monell Center now allows scientists to identify the complete set of genes in any type of taste receptor cell. The technology provides taste researchers with a treasure trove of information that will help identify precisely how each type of taste receptor cell carries out its specific function.
"The taste system is in many ways the gatekeeper of nutrition," said Monell molecular biologist Sunil Sukumaran, PhD, who co-led the research. "Our study deepens knowledge of how taste cells work, with the potential to help devise ways to manipulate taste and promote healthy eating."
Taste buds contain between 50 and 100 tightly-packed cells and are surprisingly diverse. Each of these onion-shaped sensory structures contains several different types and subtypes of taste receptor cells, each with a specialized function. Scientists know that some so-called 'type II' taste cells respond to sweet or umami (savory) taste compounds, while other type II subtypes respond to bitter. Likewise, 'type III' cells can respond to salty or sour or sometimes both.
But, much remains to be learned about how these different cells recognize and translate information from a given taste molecule and then activate a nerve to carry this information to the brain.
By identifying the genes in any given taste cell, scientists can pinpoint the unique molecular components needed for that cell to carry out its particular function. However, due to technical issues, previous studies could provide only limited information on the specific molecular components associated with each specific taste cell type.
In the new study, published online in the open-access journal Scientific Reports, the Monell researchers first identified individual type II umami/sweet sensing cells using one technique and type III salty/sour cells with a second method. They then used a powerful genetic technology known as RNA-seq on the single cells to identify all the genes expressed in each of these taste cell types.
"The genes that a taste cell expresses define essentially every aspect of that cell's behavior, from how it will respond to taste stimuli to how it will communicate with other cells," said study co-leader Brian Lewandowski, PhD, a Monell physiologist. "By identifying the full set of genes in each of these two cell types, we provide detailed information that taste scientists can use to help understand how these cells work, both when healthy and also when affected by damage or disease."
As an example, the current study identified and provided insight into the function of several genes, not previously known to be involved in the taste system, related to cell motility.
The type III salty/sour taste cells were identified using a method called calcium imaging, marking the first time that RNA-seq has been successfully used on single cells characterized using this technique. Because calcium imaging is widely used to identify cells in tissues throughout the body, researchers studying other systems will be able to benefit from the published methodology.
With regard to taste, the findings will allow researchers to create a complete map of all the genes expressed in every type of taste cell within a taste bud, leading to a better understanding of how the taste system works.
"We still have much to learn about salt, sour, and fat taste detection," said Sukumaran. "We now can formulate more informed hypotheses to answer these open questions and better understand how diet, age and numerous other factors influence the taste system."
More information: Sunil K. Sukumaran et al, Whole transcriptome profiling of taste bud cells, Scientific Reports (2017). DOI: 10.1038/s41598-017-07746-z
Journal information: Scientific Reports
Provided by Monell Chemical Senses Center