Virus's multipronged attack on bacteria points to new ways to fight infection
Researchers have uncovered how a virus uses a two-pronged attack against a single protein to kill bacteria.
The international group, led by scientists from the MRC Centre for Molecular Bacteriology & Infection at Imperial College London, believe their work could ultimately lead to new ways to combat bacterial infections.
The team made the discovery while studying a type of virus known as a T7 bacteriophage, which infects E. coli, the bacterium which can cause food poisoning.
The virus, which is hundreds of times smaller than a bacterium, selectively targets E.coli, hijacking the bacterium's biological functions to churn out more copies of itself.
Previously, a group led by Professor Ramesh Wigneshweraraj from the Department of Medicine and Professor Steve Matthews from the Department of Life Sciences, discovered that this virus attacks its bacterial host by interfering with E.coli's ability to switch on its genes.
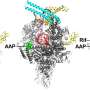
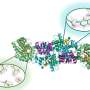
They showed that the virus produces a protein called Gp2 which targets a crucial enzyme in the bacterium known as RNA polymerase (RNAP).
This enzyme is used by bacteria to transcribe their genetic code to strands of RNA, needed for making proteins.
RNAP is already a target for some antibiotics like rifamycins, used to treat tuberculosis and leprosy, which kill bacteria by binding to the enzyme and disrupting its ability to make RNA.
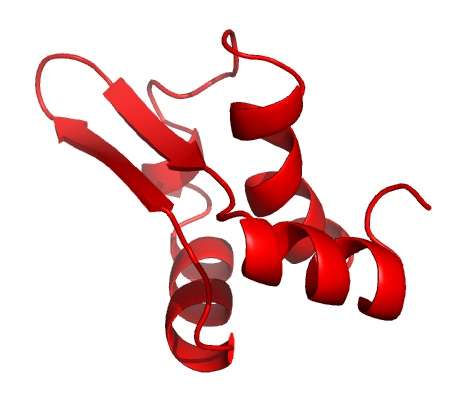
In the latest study, published in the journal Nucleic Acid Research, the international group, which involved researchers from the US, Russia and Israel, has shown that the virus also uses a second protein to target RNAP – forming a two-pronged attack against the same target in E.coli.
Doubling up
"These viruses are quite efficient killers. They don't have resources to spare, so typically they will develop one protein to kill one process," explained Professor Wigneshweraraj. "The novelty of our new research is that we have discovered that one virus has evolved two entirely different ways to attack the same bacterial target."
The group thinks that the first protein (Gp2) can only inhibit free forms of the enzyme – not actively transcribing DNA – in the bacterial cell during the early stages of infection.
However, later on in the attack, more RNAP becomes available. The virus produces the second protein (Gp5.7) to target the remaining enzymes and finish the job, resulting in a finely tuned process to kill the bacterium.
Professor Wigneshweraraj says that while the study is fundamental research and is a long way from being used in the clinic, the pharmaceutical industry could use the research to revisit existing targets in the bacterial cell for intervention.
"Our view is that by studying these proteins we can get truly new inspiration to hit on new ways to fight bacterial infections. If we can mimic the processes that viruses use, we may be able to create new forms of treatment – against proven antibiotic targets in bacteria," said Professor Wigneshweraraj.
The researchers say the next step is to focus more on how Gp5.7 binds to RNAP and to work out the exact mechanism by which the viruses take down the bacteria.
More information: Aline Tabib-Salazar et al. Full shut-off of Escherichia coli RNA-polymerase by T7 phage requires a small phage-encoded DNA-binding protein, Nucleic Acids Research (2017). DOI: 10.1093/nar/gkx370
Journal information: Nucleic Acids Research , Nucleic Acid Research
Provided by Imperial College London