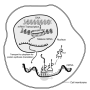
Quality control inside the cell
The ability to dispose of proteins that are either aberrant or (in the worst case) toxic is fundamental to a cell's survival. Researchers from Charité - Universitätsmedizin Berlin have been able to demonstrate the manner in which two specific proteins recognize defective messenger RNAs (molecules that carry the 'assembly instructions' for protein synthesis) and trigger their destruction. Results from this study have been published in the current issue of the journal Nature Communications.
During protein synthesis, the genetic information stored in a gene's DNA is translated into proteins. This process takes place inside veritable macromolecular machines known as ribosomes, and starts by transcribing genetic information from a cell's DNA into transportable units known as messenger RNAs (mRNAs). These units, which contain detailed instructions for the synthesis of specific proteins, are then read and translated by ribosomes into proteins. Defective mRNAs will result in aberrant, potentially harmful proteins; an efficient process for recognizing and disposing of such mRNAs is therefore essential.
As part of their study, researchers led by Dr. Tarek Hilal from Charité's Institute of Medical Physics and Biophysics studied mRNAs that lack 'stop codons' - genetic stop signs that signal the end of protein synthesis. Attempts to decode and translate such 'nonstop-mRNAs' leads to a complete stalling of the ribosomal machinery, resulting in effectively blocking continued protein synthesis. Using cryo-electron microscopy to study the structure of such ribosome-mRNA complexes, the researchers were able to show the manner in which special rescue proteins (Dom34 and Hbs1) recognize such stalled ribosomes, thereby initiating the splitting of the arrested complex and the degradation of the faulty mRNA. The rescue proteins recognize arrested ribosomes by detecting, and binding to, conserved locations normally occupied by mRNA. This direct competition-based approach ensures that only ribosomes with aberrant mRNAs are targeted.
"Research into the effects of aberrant mRNAs and the consequences of inadequate degradation is becoming increasingly significant," says Dr. Hilal. He adds: "Aberrant mRNAs have been found to be particularly common in patients with neurodegenerative disorders such as amyotrophic lateral sclerosis (ALS). Gaining an understanding of the relevant cellular control mechanisms on a molecular level may help us to develop new treatment approaches."
More information: Tarek Hilal et al, Structural insights into ribosomal rescue by Dom34 and Hbs1 at near-atomic resolution, Nature Communications (2016). DOI: 10.1038/ncomms13521
Journal information: Nature Communications
Provided by Charité - Universitätsmedizin Berlin