Discovery of a fundamental limit to the evolution of the genetic code
A study performed at IRB Barcelona offers an explanation as to why the genetic code stopped growing 3,000 million years ago. This is attributed to the structure of transfer RNAs—the key molecules in the translation of genes into proteins. The genetic code is limited to 20 amino acids—the building blocks of proteins—the maximum number that prevents systematic mutations, which are fatal for life. The discovery could have applications in synthetic biology.
Nature is constantly evolving—its limits determined only by variations that threaten the viability of species. Research into the origin and expansion of the genetic code are fundamental to explain the evolution of life. In Science Advances, a team of biologists specialised in this field explain a limitation that put the brakes on the further development of the genetic code, which is the universal set of rules that all organisms on Earth use to translate genetic sequences of nucleic acids (DNA and RNA) into the amino acid sequences that comprise the proteins that undertake cell functions.
Headed by ICREA researcher Lluís Ribas de Pouplana at the Institute for Research in Biomedicine (IRB Barcelona) and in collaboration with Fyodor A. Kondrashov, at the Centre for Genomic Regulation (CRG) and Modesto Orozco, from IRB Barcelona, the team of scientists has demonstrated that the genetic code evolved to include a maximum of 20 amino acids and that it was unable to grow further because of a functional limitation of transfer RNAs—the molecules that serve as interpreters between the language of genes and that of proteins. This halt in the increase in the complexity of life happened more than 3,000 million years ago, before the separate evolution of bacteria, eukaryotes and archaebacteria, as all organisms use the same code to produce proteins from genetic information.
The authors of the study explain that the machinery that translates genes into proteins is unable to recognise more than 20 amino acids because it would confuse them, which would lead to constant mutations in proteins and thus the erroneous translation of genetic information "with catastrophic consequences", in Ribas' words. "Protein synthesis based on the genetic code is the decisive feature of biological systems and it is crucial to ensure faithful translation of information," says the researcher.
A limitation imposed by shape
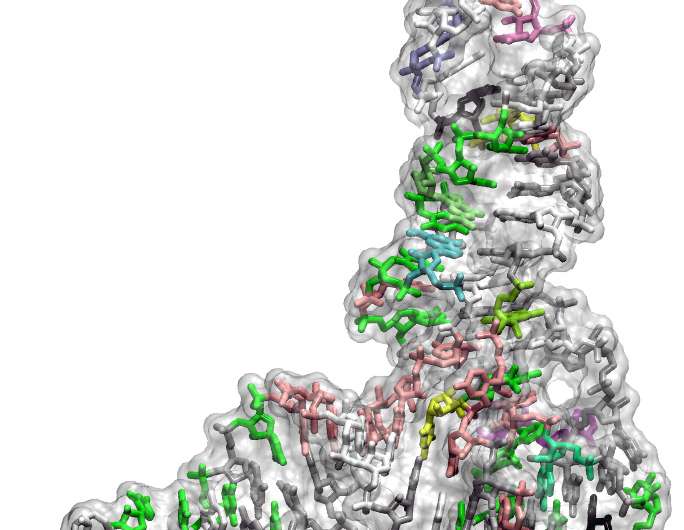
Saturation of the genetic code has its origin in transfer RNAs (tRNAs), the molecules responsible for recognising genetic information and carrying the corresponding amino acid to the ribosome, the place where chain of amino acids are made into proteins following the information encoded in a given gene. However, the cavity of the ribosome into which the tRNAs have to fit means that these molecules have to adopt an L-shape, and there is very little possibility of variation between them. "It would have been to the system's benefit to have made new amino acids because, in fact, we use more than the 20 amino acids we have, but the additional ones are incorporated through very complicated pathways that are not connected to the genetic code. And there came a point when Nature was unable to create new tRNAs that differed sufficiently from those already available without causing a problem with the identification of the correct amino acid. And this happened when 20 amino acids were reached," explains Ribas.
Application in synthetic biology
One of the goals of synthetic biology is to increase the genetic code and to modify it to build proteins with different amino acids in order to achieve novel functions. For this purpose, researchers use organisms such as bacteria in highly controlled conditions to make proteins of given characteristics. "But this is really difficult to do and our work demonstrates that the conflict of identify between synthetic tRNAs designed in the lab and existing tRNAs has to be avoided if we are to achieve more effective biotechnological systems," concludes the researcher.
More information: A. Saint-Leger et al. Saturation of recognition elements blocks evolution of new tRNA identities, Science Advances (2016). DOI: 10.1126/sciadv.1501860
Journal information: Science Advances