Insights into genomic instability during the early stages of embryonic cell development
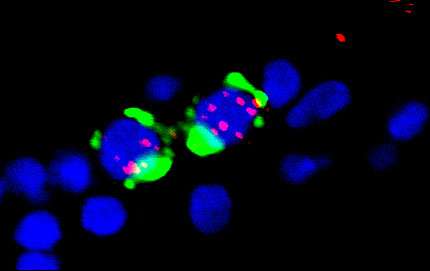
The environment within a cell is a paradoxical balance of order and chaos. Proteins, DNA and countless other biological molecules tumble together in a wildly turbulent system that somehow holds together to engender life.
In the earliest stages of an organism's existence, however, cells hazard an even greater level of disorder—one that threatens to tear their DNA apart. The phenomenon is the focus of research by doctoral student Melina Butuci and her mentor, Matthew Michael, associate professor of biological sciences.
Trouble in the transition
Just after two gametes (a sperm cell and an egg cell) join to form a zygote, the new, single cell remains under the control of leftover proteins from the original egg cell. As the zygote begins to divide—first forming two cells, then four, and so on—to form an embryo, the residual maternal proteins dwindle. The embryonic cells, recognizing their waning resources, suddenly activate their genome and begin rapidly reading their own genetic instructions, transcribing the code so they can produce the proteins they need for themselves.
This rapid transition, called zygotic genome activation, or ZGA, is necessary for the cell's survival, but it paradoxically leads to a great deal of instability and potentially life-threatening damage to the cells' DNA.
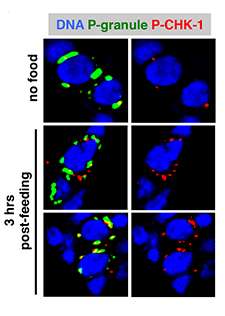
Finding answers in a worm
Studying primitive cells called germ cells from a small roundworm known as Caenorhabditis elegans, Butuci and Michael found that ZGA-related genomic instability and DNA damage stems not from the increased transcription of genes once the system is in motion, but rather from the sudden change—when the zygotic genome's "switch" is suddenly flipped on.
"We saw that the transcription initiation leads to the [DNA] breaks," Butuci said. "This could happen because the transcription might be so abrupt and a large amount of it happens at the same time."
Because DNA is tightly packed into chromosomes within cells, it first must be unraveled and opened up for gene-reading machinery to access it. This makes it inherently less stable and prone to damage under the best of circumstances. During ZGA, however, an enormous number of genes are being accessed very suddenly and all at once, spreading instability throughout the genome as the system jolts into motion.
In addition, Michael and Butuci found that an enzyme called topoisomerase II is involved in opening up the DNA during ZGA. Topoisomerase II works by severing DNA across the double helix, and while this opens the DNA up for transcription, the scientists think it also might leave it vulnerable to permanent damage and be a source of genomic instability. Butuci cautioned that this hypothesis is unproven and further research to better understand the phenomenon is underway.
A "bizarre" evolutionary trade-off
Despite the seemingly life-threatening genomic instability arising during embryonic development, the cells survive. The researchers found that highly robust repair mechanisms—much more accurate and efficient than those found in more mature cells—reverse the DNA damage that arises from ZGA. This restores the balance between the need for rapid activation of genes and the risk of permanent, lethal DNA breaks.
"The general idea is that when transcription is low, chromosomes tend to be quite condensed, and thus transcription is difficult. Now, when a signal is received to activate transcription, it may be that cells use DNA breaks as a way to quickly decondense the DNA," Michael explained. "So, what cells are doing is trading the risk of mutation (because of the break) for speed in activating gene expression. It's easy to imagine that a rapid gene expression response is [evolutionarily] very advantageous, and as long as repair is efficient, then this seems like a reasonable trade."
The research, which is part of Butuci's doctoral studies, continues as the scientists further unravel this genomic puzzle. "As a scientist, I am extremely intrigued to understand more about this mechanism and the evolutionary path that just seems so bizarre at first glance, but surely has arisen with a great purpose," Butuci said.
The study appears in the July 6 issue of Developmental Cell.
More information: "Zygotic Genome Activation Triggers Chromosome Damage and Checkpoint Signaling in C. elegans Primordial Germ Cells." DOI: 10.1016/j.devcel.2015.04.019
Journal information: Developmental Cell
Provided by University of Southern California