Fusion protein controls design of photosynthesis platform
Chloroplasts are the solar cells of plants and green algae. In a process called photosynthesis, light energy is used to produce biochemical energy and the oxygen we breathe. Thus, photosynthesis is one of the most important biological processes on the planet. A central part of photosynthesis takes place in a specialized structure within chloroplasts, the thylakoid membrane system. Despite its apparent important function, until now it was not clear how this specialized internal membrane system is actually formed. In a collaborative project, researchers at Johannes Gutenberg University Mainz (JGU) in Germany have now identified how this membrane is generated. According to their findings, a protein called IM30 plays a major role by triggering the fusion of internal membranes. The study elucidating the role of IM30 involved biologists, chemists, biochemists, and biophysicists at Mainz University and the Max Planck Institute for Polymer Research. Their results have recently been published in the journal Nature Communications.
Chloroplasts are organelles found in higher plants and green algae. They contain an internal membrane system, so-called thylakoid membranes, where the key processes of photosynthesis take place. "A detailed understanding of photosynthesis and the associated molecular processes is essential to properly comprehend life on our planet," emphasized Professor Dirk Schneider of the Institute of Pharmaceutical Sciences and Biochemistry at JGU, who coordinated the study. "Despite the significance of the process, we know almost nothing about how these special membranes are formed and maintained." It had not previously been possible to identify a single fusion-mediating protein in photosynthetic cells, even though it was perfectly clear that such proteins have to be involved in the development of thylakoid membranes.
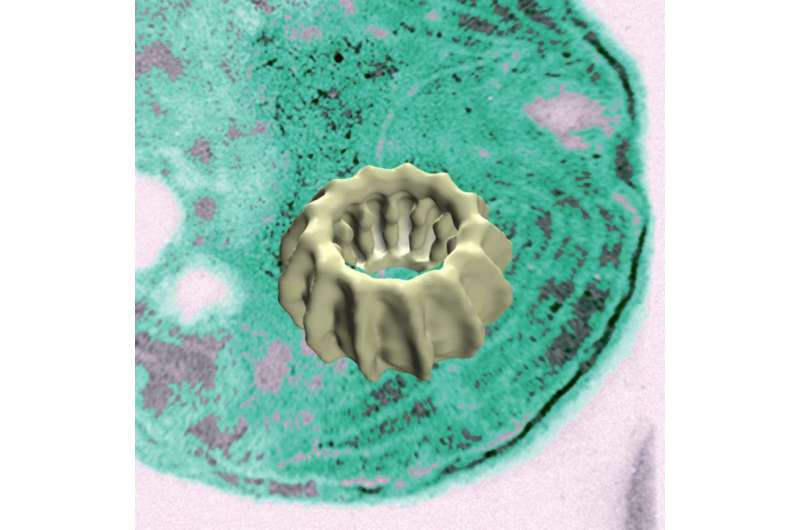
With this in mind, the Mainz-based research team isolated and investigated the protein IM30 from a blue-green alga, which might be classified as a "free-living chloroplast." IM30 - the "IM" stands for "internal membrane" while 30 is its atomic mass (30 kilodaltons) - was first described in the mid-1990s and it was demonstrated that it binds to internal membranes. Thanks to the combined expertise of the teams headed by Professor Dirk Schneider, Professor Jürgen Markl of the JGU Institute of Zoology, and Professor Tobias Weidner of the Max Planck Institute for Polymer Research it has now emerged that IM30 forms a ring structure that specifically interacts with phospholipids of the membranes. "This binding alters the membrane structure and under certain conditions can lead to membrane fusion," explained Schneider. In absence of IM30, thylakoid membranes are noticeably deteriorated, which can subsequently lead to loss of cell viability. The IM30 fusion protein provides a starting point for future research, unraveling new types of membrane fusion mechanisms in chloroplasts and blue-green algae.
The interdisciplinary research project was primarily undertaken by doctoral candidates at the Max Planck Graduate Center (MPGC). The MPGC was founded in June 2009 to support joint projects and shared doctorates at Johannes Gutenberg University Mainz and the Max Planck Institutes for Polymer Research and for Chemistry, both of which are based in Mainz.
More information: Raoul Hennig et al., IM30 triggers membrane fusion in cyanobacteria and chloroplasts, Nature Communications, 8 May 2015. DOI: 10.1038/ncomms8018
Journal information: Nature Communications
Provided by Universitaet Mainz