New technique to chart protein networks in living cells
A new approach for studying the behaviour of proteins in living cells has been developed by an interdisciplinary team of biologists and physicists in the Cell Biology and Biophysics Unit, the Ellenberg Laboratory and the Advanced Light Microscopy Facility at the European Molecular Biology Laboratory in Heidelberg.
Described in a new study, published today in Nature Biotechnology, the approach allows scientists for the first time to follow the protein networks that drive a biological process in real time.
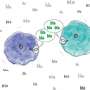
Which proteins interact with each other and where they meet within cells is of huge interest to scientists because it reveals the state and activity of the molecular machinery that drives the most fundamental functions of life such as the ability of cells to divide. The new technique will also be useful for scientists to investigate the mechanisms of disease and for pharmaceutical companies to explore new drug targets.
At the heart of this technology is a method called fluorescence correlation spectroscopy (FCS). Originally developed in the 1970s, FCS enables scientists to track individual proteins inside living cells once they have been fused to a fluorescent marker. FCS can measure where proteins move and when they meet, in a similar way to how Google can track people in traffic jams with their smartphone GPS signals.
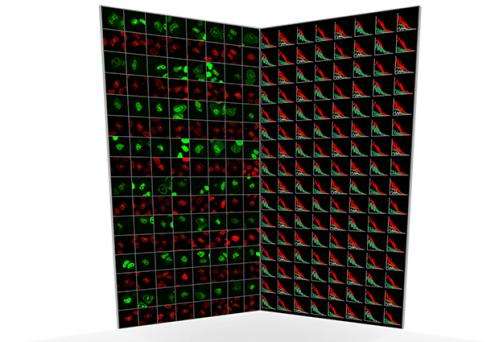
Although such data would be tremendously valuable to have for all the proteins within a cell, until now FCS microscopes have been very cumbersome to use and the data difficult to interpret. The new development at EMBL automates the whole process of measuring protein behaviour and analysing the very large amounts of single molecule data.
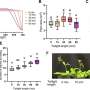
"Our approach enables huge time savings" explains Malte Wachsmuth, of EMBL, who co-authored the study. "Whereas previously a scientist might spend a day observing one protein in a few cells, we can now acquire data from tens of proteins in thousands of cells fully automatically."
"Because we have made FCS into a high throughput method, we can acquire data from many different proteins, which is key to studying biological networks that typically consist of tens to hundreds of components" says Jan Ellenberg.
More information: Wachsmuth M. et al. High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Advanced online publication in Nature Biotechnology on the 16 March, 2015. DOI: 10.1038/NBT.3146
Journal information: Nature Biotechnology
Provided by European Molecular Biology Laboratory