Counting small RNA in disease-causing organisms
Small molecules of RNA (tens to hundreds of nucleotides in length) play a key regulatory role in bacteria. Due to their small size, directly measuring the number of small RNA (sRNA) present in a single bacterium has proven so far to be an impossible task. Standard methods of measuring the number of specific nucleic acid molecules present in a single cell suffer from too much background and false positives when scientists attempt to image short targets. In research featured on the cover of the journal Analytical Chemistry, Los Alamos researchers demonstrated improved technical methods capable of directly counting small RNA molecules in pathogenic (disease-causing) bacteria.
The ability to quantify the expression level changes of sRNA in single cells as a function of external stimuli provides important information to understand the role of sRNA in cellular regulatory networks and disease. The scientists found that only a small fraction of either Yersinia pestis (causative agent for the plague) or Yersinia pseudotuberculosis bacteria express two sRNAs thought to be important in pathogenesis, with the copy number typically between 0-10 transcripts per bacteria. The relatively low sRNA copy number in the bacterial populations raises the question of how many sRNAs are required for regulatory control over messenger RNA (mRNA) and protein populations.
The team learned that the change in the distribution of sRNAs per bacterium for a situation that mimics infection of a warm-blooded host could be described through a bursting model of gene transcription, where sRNA are produced for a discrete period of time. The numbers of these sRNAs increased upon a temperature shift from 25 to 37 °C, corresponding to a change from Y. pestis infection conditions of the flea host to a human host during infection. These results suggest that the sRNAs play a role in pathogenesis. The team's modeling suggests that sRNA copy numbers are controlled by a change in the on-rate of the gene under the human host infection conditions.
Research achievements
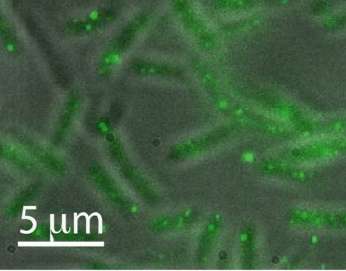
The team directly visualized single short (approximately 200 nucleotide) nucleic acid targets in bacteria by neutralizing the fluorescence of unbound dye probes. They used an automated, multi-color wide-field microscope and data analysis package to analyze the statistics of sRNA expression in thousands of individual bacteria. The new technique reduced the number of false positives, which improved the accuracy of the count statistics, and it significantly reduced the image processing time. The team performed modeling of the sRNA increase under simulated conditions of infection in a human host, finding a bursting model of gene transcription well described the observed sRNA upregulation.
Journal information: Analytical Chemistry
Provided by Los Alamos National Laboratory