How yeast formations got started
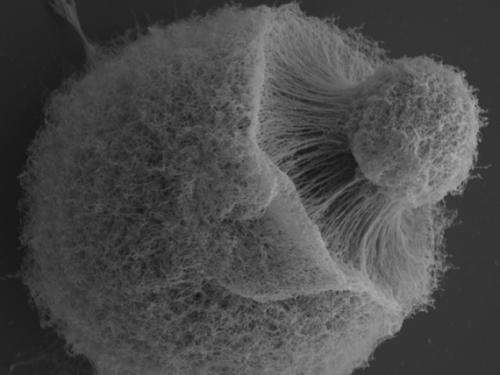
Researchers conducted a comparative analysis of nearly 60 fungal genomes to determine the genetic traits that enabled the convergent evolution of yeasts.
The analyses suggest that a conserved zing-finger transcription factor family allowed yeasts to become the dominant form in multiple fungal clades. This "parallel deployment" is not consistent with the classical notion of convergent evolution.
Yeasts are found in multiple fungal clades and one of the most familiar is Saccharomyces cerevisiae, used both in food preparation and as a model organism for developing biofuels. While many yeasts have a unicellular form, several yeasts can also take the form of filamentous fungi. Despite their divergent lineages, yeasts have many phenotypic and metabolic similarities. To learn more about the convergent evolution of yeasts, a team including researchers at the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility, and several longtime DOE JGI collaborators, conducted a comparative analysis of nearly 60 fungal genomes. Their findings appeared ahead online July 18, 2014 in Nature Communications.
Many of the genomes used in the study were sequenced and analyzed at the DOE JGI. For the study, the team developed a computational pipeline called COMPARE or comparative phylogenomic analysis of trait evolution in order to identify genes that had been inherited from a shared ancestor (orthologous genes) and determine gene duplications or losses on charted phylogeny trees. The team pegged the origin for the potential of yeast-like growth at some 770 million years ago. They also noted several gene classes that were conserved, likely due to their roles in essential processes such as DNA replication and multicellular growth.
"We suggest that the genetic toolkit for yeast-like growth has been added on top of the eukaryote cell division programme early in fungal evolution and has been widely conserved throughout fungal evolution," the team wrote. "This finding explains the widespread occurrence of yeasts and dimorphic fungi with remarkably similar yeast phases among phylogenetically diverse fungal groups."
In particular, they identified several genes across the species related to a single transcription factor (TF) family called Zn-cluster TFs. This family, they noted, regulates processes such as the ability to change forms from yeasts to filamentous fungi. They suggest that this mechanism may be involved in the convergent evolution, or the process in which different organisms that not closely related will independently evolve similar traits that serve similar functions, of the yeast phenotype.
More information: Nagy LG et al. Latent homology and convergent regulatory evolution underlies the repeated emergence of yeasts. Nat. Commun. 2014 July 18;(5):4471. DOI: 10.1038/ncomms5471
Journal information: Nature Communications
Provided by DOE/Joint Genome Institute