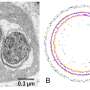
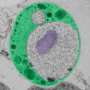
Scientists study how complexity developed from simple cell
(Phys.org) —Consider this a matter of scrambling down the family tree to its roots. Really old roots. Or perhaps it's more like blowing the dust off the family album—the human album—and opening to the first pages billions of years ago.
Naomi Ward, an associate professor in the University of Wyoming Department of Molecular Biology, is the senior author on a paper recently published in the Proceedings of the National Academy of Sciences (PNAS).
The research examines how simple bacterial cells could have made the transition to more complex cells, leading to plants, animals and humans.
The paper, titled "Spatially segregated transcription and translation in cells of the endomembrane-containing bacterium Gemmata obscuriglobus," was published online this week. It describes research supported by a grant from the National Science Foundation (NSF).
Ekaterina Gottshall, a graduate student in the Molecular and Cellular Life Sciences Ph.D. program, is first author on the paper and main contributor to the experimental work. Other authors are assistant professor Jay Gatlin, also in molecular biology, and Corrine Seebart, an assistant research scientist in Ward's group.
Ward's version of genealogy looks at how simple bacterial cells, which do not have the nuclear membrane that separates transcription and translation (the reading of DNA instructions to make protein), could have evolved into eukaryotic cells (plants, animals, humans), in which transcription and translation occur in separate locations.
This evolutionary step was an important part of developing greater cell complexity in ancient eukaryotic cells.
"This is usually considered to be a very fundamental way in which bacterial cells differ from our cells," Ward says. "However, cells of Gemmata obscuriglobus (the bacterium they studied) have complex internal membranes, making them look superficially like eukaryotic cells."
Gottshall wanted to know if transcription and translation could occur in different places in the cell just like in a eukaryote cell.
"We asked this question because the way in which complex eukaryotic cells evolved from a simpler ancestor is not completely understood, and we thought that studying this question in Gemmata might shed some light on that problem," Gottshall says.
It is generally thought that two of the major membrane-bound compartments in animal and plant cells—mitochondria, the power plants of the cell, and chloroplasts, where photosynthesis occurs—were formed when ancient bacteria took up residence in an ancient proto-eukaryotic cell.
Some estimates place the move-in date around 1.8 billion years ago. Bacterial microfossils first appear about 3.5 billion years ago.
Ward and her research group found a substantial amount of G. obscuriglobus translation does occur in a different place from transcription, as is found in eukaryotic cells.
"Although this is not the first time this has been reported for bacteria, it is the first time it has been reported for such a complex bacterial cell," Ward says. "Although we don't know whether this uncoupled gene expression in Gemmata arose in the same way it did in the ancient eukaryotic cell, it shows us one possible way in which it might have been organized."
Ward participated in the recent art-science collaborative experiment (The Ucross-Pollination Experiment), organized by UW Philosophy Professor Jeff Lockwood. She collaborated with Philosophy Professor H.L. Hix to explore form in poetry and science, and one of the products was a poem based on the PNAS paper.
More information: Ekaterina Y. Gottshall, Corrine Seebart, Jesse C. Gatlin, and Naomi L. Ward. "Spatially segregated transcription and translation in cells of the endomembrane-containing bacterium Gemmata obscuriglobus." PNAS 2014 ; published ahead of print July 14, 2014, DOI: 10.1073/pnas.1409187111
Journal information: Proceedings of the National Academy of Sciences
Provided by University of Wyoming