Newly discovered mechanism could regulate gene activity
Many bacterial species have genes called mraZ and mraW, which are located in a cluster of genes that regulate cell division and cell wall synthesis. Despite the prevalence of these two genes, very little is known about their functions. This study reveals that mraZ and mraW work in opposing ways to control cell growth and division, and that mraZ encodes a transcription factor that binds DNA to potentially regulate the activity of many other genes.
The findings shed light on a newly discovered mechanism that many bacteria may potentially use to adapt to suboptimal environments or growth conditions. Thus, this research contributes to the mission of the Department of Energy's Biological and Environmental Research program, which aims to define the principles that guide the translation of the genetic code into functional proteins and the metabolic/regulatory networks underlying the systems biology of plants and microbes as they respond to and modify their environments.
To determine the functions of mraZ and mraW, researchers from the Department of Energy's Pacific Northwest National Laboratory, EMSL and the University of Texas Medical School at Houston used molecular biology tools to enhance gene transcription as well as inactivate the two genes in Escherichia coli (E. coli). They performed transcriptome analysis and used bioinformatics resources at EMSL, the Environmental Molecular Sciences Laboratory, a DOE national scientific user facility.
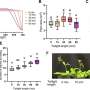
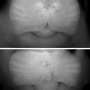
They found that high levels of the MraZ protein inhibited cell division and killed the cells, and excess MraZ was more toxic when the cells were provided with only the minimum nutrients possible for colony growth rather than all of the necessary nutrients for robust growth. Moreover, MraZ toxicity increased when the bacteria were genetically manipulated to lack the MraW protein, whereas MraZ toxicity decreased when MraZ and MraW were simultaneously overproduced. These findings suggest MraW counteracts the toxic effects of MraZ, and the two proteins work in opposing ways to regulate cell division and growth.
The researchers also found that MraZ binds DNA to potentially regulate the activity of many other genes. In support of this idea, loss of MraZ affected the activity of about 2% of genes in the E. coli genome, and overproduction of this protein affected the activity of nearly one-quarter of all E. coli genes, including those involved in cell division, cell wall synthesis and metabolism. Taken together, the results suggest that MraZ may inhibit cell wall synthesis and cell division under conditions of nutritional stress to maintain the proper balance between growth and nutrient availability. Because mraZ and mraW are highly conserved, these novel insights into their functions will likely translate to other bacterial species.
More information: Eraso, J.M., Markillie, L.M., Mitchell, H.D., Taylor, R.C., Orr, G., and Margolin, W. "The highly conserved MraZ protein is a transcriptional regulator in Escherichia coli." Journal of Bacteriology 196, (2014). DOI: 10.1128/JB.01370-13
Journal information: Journal of Bacteriology
Provided by Environmental Molecular Sciences Laboratory