New method reveals single protein interaction key to embryonic stem cell differentiation
Proteins are responsible for the vast majority of the cellular functions that shape life, but like guests at a crowded dinner party, they interact transiently and in complex networks, making it difficult to determine which specific interactions are most important.
Now, researchers from the University of Chicago have pioneered a new technique to simplify the study of protein networks and identify the importance of individual protein interactions. By designing synthetic proteins that can only interact with a pre-determined partner, and introducing them into cells, the team revealed a key interaction that regulates the ability of embryonic stem cells to change into other cell types. They describe their findings June 5 in Molecular Cell.
"Our work suggests that the apparent complexity of protein networks is deceiving, and that a circuit involving a small number of proteins might control each cellular function," said senior author Shohei Koide, PhD, professor of biochemistry & molecular biophysics at the University of Chicago.
For a cell to perform biological functions and respond to the environment, proteins must interact with one another in immensely complex networks, which when diagrammed can resemble a subway map out of a nightmare. These networks have traditionally been studied by removing a protein of interest through genetic engineering and observing whether the removal destroys the function of interest or not. However, this does not provide information on the importance of specific protein-to-protein interactions.
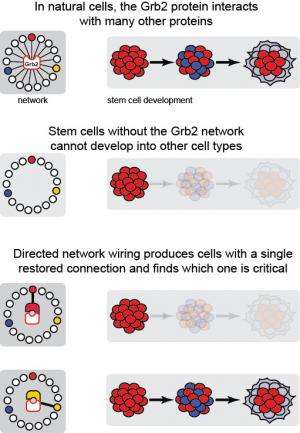
To approach this challenge, Koide and his team pioneered a new technique that they dub "directed network wiring." Studying mouse embryonic stem cells, they removed Grb2, a protein essential to the ability of the stem cell to transform into other cell types, from the cells. The researchers then designed synthetic versions of Grb2 that could only interact with one protein from a pool of dozens that normal Grb2 is known to network with. The team then introduced these synthetic proteins back into the cell to see which specific interactions would restore the stem cell's transformative abilities.
"The name, 'directed network wiring,' comes from the fact that we create minimalist networks," Koide said. "We first remove all communication lines associated with a protein of interest and add back a single line. It is analysis by addition."
Despite the complexity of the protein network associated with stem cell development, the team discovered that restoring only one interaction—between Grb2 and a protein known as Ptpn11/Shp2 phosphatase—was enough to allow stem cells to again change into other cell types.
"We were really surprised to find that consolidating many interactions down to a single particular connection for the protein was sufficient to support development of the cells to the next stage, which involves many complicated processes," Koide said. "Our results show that signals travel discrete and simple routes in the cell."
Koide and his team are now working on streamlining directed network wiring and applying it to other areas of study such as cancer. With the ability to dramatically simplify how scientists study protein interaction networks, they hope to open the door to new research areas and therapeutic approaches.
"We can now design synthetic proteins that are far more sophisticated than natural ones, and use such super-performance proteins toward advancing science and medicine," he said.
More information: "Directed network wiring identifies a key protein interaction in embryonic stem cell differentiation," Molecular Cell.
Journal information: Molecular Cell
Provided by University of Chicago Medical Center