Research reveals evolution of cells' signaling networks in diverse organisms
(Phys.org) —Cells use protein-signaling networks to process information from their surroundings and respond to constantly changing environments. This includes information about the presence or absence of vital nutrients as well as the presence of other cells. Signaling networks control the decisions that cells make in response to these conditions.
In a paper just published in the influential Proceedings of the National Academy of Sciences, researchers from the University of Kansas shed light on protein-signaling networks used by bacterial cells and why they are so different from the more complex networks found in eukaryotes, such as human beings.
"Our research focuses on elucidating the general principles that underlie how signaling networks work in a variety of organisms," said Eric Deeds, assistant professor at KU's Center for Bioinformatics and Department of Molecular Biosciences. "We're helping people understand why they observe the types of networks that they do in different cells."
According to Deeds, a better understanding of information processing inside human cells will lead to more rational approaches to targeting complex diseases like cancer.
"In bacterial systems, our work could help lead to the development of novel antibiotic strategies as well as the engineering of new information processing capabilities in bacterial cells," he said.
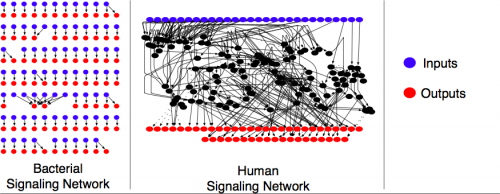
Human cells possess very complex networks with many interconnected pathways. Bacteria, on the other hand, utilize very simple networks in which most pathways are completely isolated from one another.
"It's like the contrast between a telephone wire and a complicated computer chip," said Deeds. "A telephone wire connects one person to another directly and simply transmits the signals from one end to the other. This is like the bacterial signaling pathways, which have a simple topology directly connecting inputs to outputs. In contrast, a computer chip is highly interconnected, with many downstream elements responding to any given input. This kind of integrated architecture allows a computer to perform complex signal-processing tasks. The architecture of human signaling networks is similar to that of a computer chip, with a much more complicated topology and a much richer possible set of input-output relationships."
The paper in PNAS authored by Deeds and KU graduate research assistant Michael Rowland shows that this global difference in network structure ultimately derives from the properties of the atomic "building blocks" from which the networks themselves are constructed.
"In particular, the enzymes used in bacterial-signaling networks are often bifunctional, in that they catalyze both the modification and de-modification of their protein substrates," Deeds said.
The research showed that these bifunctional enzymes become progressively less efficient as they work on larger numbers of substrates. This prevents them from evolving the extensive levels of "crosstalk" that characterize eukaryotic networks. In contrast, the enzymes employed by human cells are "monofunctional," leading to "remarkably different" global architectures.
"Monofunctional enzymes basically represent a 'division of labor,'" Deeds said. "There is one protein that performs the phosphorylation step—called the kinase—and another that performs the dephosphorylation step, called the phosphatase. Our work has shown that dividing things up like this allows human networks to achieve high levels of crosstalk, producing the complex integrated architectures that perform decision-making tasks for our cells. Bacterial networks, which use a single enzyme to perform both those tasks, simply can't exhibit high levels of crosstalk."
Moving ahead, Deeds' work aims to achieve a firmer understanding of the crosstalk that characterizes complex protein signaling in humans.
"Although it's clear that human networks can and do have massive amounts of crosstalk, we currently don't understand exactly what these systems are doing," said Deeds. "Fully answering that question is a major area of research both for my lab and for a large number of other labs around the world."
Journal information: Proceedings of the National Academy of Sciences
Provided by University of Kansas