Scientists develop largest developmental proteomic data set for any animal

(Phys.org) —Now that the human genome is sequenced, University of Notre Dame researchers are focusing on the study of the proteome, which is the protein content of an organism, tissue or cell. Bioanalytical chemist Norman Dovichi and molecular biologist Paul Huber have successfully tracked the changing patterns of protein expression during early development of Xenopus laevis, or African clawed frog, embryos. They have developed the largest data set on developmental proteomics for any organism, and have included the single-cell zygote.
Their research has uncovered an unexpected amount of discordance between the levels of messenger RNA (mRNA) and its corresponding protein. Their findings are published in Scientific Reports in an article titled, "Quantitative proteomics of Xenopus laevis embryos: expression kinetics of nearly 4000 proteins during early development."
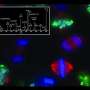
The Notre Dame team based in the Department of Chemistry and Biochemistry in the College of Science has identified and measured the levels of about 4,000 proteins, which exhibited patterns of expression that reflect key events during early Xenopus development. For example, the appearance of organ- and tissue-specific proteins, such as those found exclusively in cardiac muscle cells, accurately reflects imminent anatomical changes taking place in the embryo. The research could lead to insight into congenital birth defects that result from the misregulation of gene expression.
The research also contradicted a widely held assumption that the levels of mRNA, which encodes proteins, would be directly related to protein levels. While that was true in most cases, there were a surprisingly high number of exceptions, demonstrating that the amounts of a particular protein can be controlled by multiple mechanisms.
Because development takes place in well-defined stages outside the mother, Xenopus is a favored model. Embryogenesis can be easily monitored in real time; fate maps for organ development have been determined and major regulators of these processes have been identified and characterized, providing an abundance of tissue- and organ-specific markers to track embryo formation. Additionally, embryos develop rapidly, achieving a nearly fully developed nervous system within four days. "It's easy to manipulate the embryos to mimic certain disease states, making Xenopus extremely valuable to biologists," Huber said.
"The collaborative, ground-breaking work of Norm Dovichi, Paul Huber and their team is crucial to helping us understand the complexity of life. We are proud of this important milestone," said Greg Crawford, dean of the College of Science at the University of Notre Dame.
More information: "Quantitative proteomics of Xenopus laevis embryos: expression kinetics of nearly 4000 proteins during early development." Liangliang Sun, et al. Scientific Reports 4, Article number: 4365 DOI: 10.1038/srep04365 . Received 03 January 2014 Accepted 17 February 2014 Published 14 March 2014
Journal information: Scientific Reports
Provided by University of Notre Dame