November 27, 2013 feature
Express yourself: Scientists use genetic algorithm to design self-assembling ssDNA-grafted particles
(Phys.org) —Material design usually follows what is known as the Edisonian method, a traditional process characterized by trial-and-error discovery rather than a systematic theoretical approach. While this may be somewhat inaccurate – Edison made use of available theories and resorted to trial and error only when no adequate theory existed – a better alternative is an a priori approach in which desired properties are defined and corresponding structures designed. To that end, scientists at Columbia University – Department of Chemical Engineering and Brookhaven National Laboratory (BNL) have recently created a design approach in which colloids grafted with single-stranded DNA self-assemble into desired structures. The researchers state that their methodology can be easily generalized, is fast and highly selective, accurately reproduces the parameters relevant to four currently realized crystals but also – surprisingly – elucidates four currently unobserved structures. While the scientists acknowledge that these structures need to be experimentally validated, they are confident that their methodology has broad potential applications.
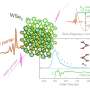
Prof. Venkat Venkatasubramanian discussed the paper that he, Prof. Sanat Kumar, Prof. Babji Srinivasan, Thi Vo from Columbia, and their co-authors, Dr. Oleg Gang and Dr. Yugang Zhang from BNL, published recently in Proceedings of the National Academy of Sciences. "Design of DNA-grafted nanoparticles is challenging because a number of experimental parameters can play a key role in self-assembly," Venkatasubramanian tells Phys.org. "Parameter space size can therefore become very large, with a large number of local minima, making it very difficult – if not impossible – to efficiently search the space in an Edisonian trial-and-error fashion." The researchers' genetic algorithm inverse design framework searches the space more systematically, making the design process more effective. (A genetic algorithm, or GA, mimics the process of natural selection.)
That being said, however, Venkatasubramanian adds that another challenge in their approach is that they require reliable forward models. "While detailed models of DNA-mediated self-assembly tend to be more accurate, they would typically require simulation times ranging from several hours to a few days, and thus would not be appropriate for coupling with the genetic algorithm for rational design." The researchers therefore chose a simpler complementary contact model (CCM) based on the predictable and well-established properties of both DNA (persistence length, rise per base pair) and DNA-NPs (number of DNA strands per particle, the hybridization behavior of sticky ends)1. "CCM has been quite successful in capturing the majority of the experimental observations," he adds, "and being fast, it allows for effective GA coupling, generating predictions of the required design parameters in a matter of minutes."
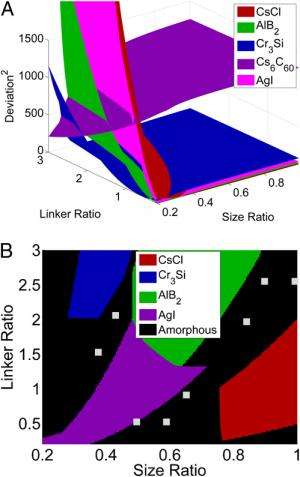
Kumar notes that experimental validation of their approach is another, and very important, challenge. "We decided to test our framework by first trying to predict nanocrystal structures that have already been observed experimentally. Next, we created a library of crystal structures – including those obtained experimentally – specifying the relevant experimental parameters, such as DNA linker ratio and the size of the DNA grafted nanoparticles. Then we ran the genetic algorithm for a desired crystal structure that has been observed experimentally, using the CCM as a forward model. We were pleased that the genetic algorithm correctly predicted the experimental parameters that were used in the formation of the observed structure."
Srinivasan, the paper's other co-author, acknowledges that currently the forward CCM model used in the GA framework has certain limitations. "It assumes complete DNA hybridization within the contact areas between two DNA-grafted particles as being the driving force behind self-assembly," he explains. "In essence, this restricts the analysis to purely attractive interaction effects. To provide a more complete enthalpic picture, we plan on building repulsive interactions between the non-complementary ssDNA linkers on the nanoparticles." Enthalpy is a measure of the total energy of a thermodynamic system.
"Furthermore," Srinivasan continues, "entropy also provides a constraint on the actual number of hybridized linkers within the nanoparticle interaction area, as well as a cost on the configuration of particles within the lattice – both of which are currently being built into the model. In addition, the current CCM does not always uniquely define a crystal structure." For example, he illustrates, the CCM cannot distinguish between the structures of CuAu (a copper/gold alloy) and CsCl (cesium chloride), since in the unit cell both crystals have the same number of nearest neighbors and particle distribution. "This gives rise to degeneracies that we plan to address by further refining the interaction types within a crystal lattice."
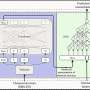
Addressing these challenges was in itself no mean feat. "Our design problem is made complicated by the complex nonlinear relationships between colloid size ratios, the number of DNA linkers on each nanoparticle and the desired crystal structure," Venkatasubramanian tells Phys.org. "This leads to a search space riddled with local minima which is difficult to explore using more conventional approaches such as trail-and-error, heuristics or mathematical programming methods." The key innovation here is the inverse design framework that effectively exploits the knowledge from the CCM (also known as a forward model) by combining it with the genetic algorithm (which is more robust to local minima), thereby resulting in a very efficient and scalable design optimization procedure. "Genetic algorithms are how nature designed complicated molecules and organisms. We're essentially creating situations where we allow the 'gene pool' of DNA-grafting parameters to evolve towards the desired – that is, 'fittest' – structure with each successive generation."
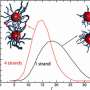
Kumar also points out that they introduced the very important analysis of cluster formation determination. "The CCM requires an initial input of crystal lattice parameters and thus assumes crystal formation given any DNA-grafting parameters. However, experimentally, there are large regions of cluster formation that result from the kinetic trapping of these particles in a metastable state during the self-assembly process. In order to prevent design falling within the boundaries of these cluster forming regimes, we performed a symmetry analysis of the model that accounts for unequal distribution of DNA linkers on particles." In other words, by introducing a cutoff on this distribution, the scientists were able to identify the presence of amorphous and cluster regions that were observed earlier in experiments. "This then allows for future modifications of the framework to incorporate an additional parameter into the fitness function," he adds, "such that it will recognize and avoid these 'bad' evolutionary paths."
Currently, by having a different objective compared to DNA hybridization, the team is including new parameters in the model to enhance the CCM with entropic contributions and repulsive interactions. "Preliminary experimental studies have shown that our refined model is in agreement with experimental results," Srinivasan and Thi Vo note, "and a thorough analysis is currently being conducted to develop a model that will uniquely specify all 230 different crystal space groups, which will then be used along with the genetic algorithm approach for crystal lattice design. Our ultimate goal is to create the model in such a way that utilizes only the parameters defining the DNA-grafted particles." The key point here is that this avoids the need for fitting constants while expanding our overall design capabilities, since the design parameters derived from the genetic algorithm framework will only involve factors that the researchers can experimentally control.
One interesting and unexpected result was that that their findings elucidated four currently unobserved structures. "Among the crystal lattice library created using the Inorganic Crystal Structure Database (ICSD), we were able to identify the parameters that could potentially lead to the formation of four new crystal structures along with those which have been observed experimentally," Srinivasan and Vo recount. "These results are promising and play a key role in the design of DNA-grafted nanomaterials." He adds that the framework is generic and can be extended to rational design of advanced materials – but with a caution that unlike molecular dynamic models, the forward path model should be computationally efficient while accounting for important aspects of the process. "We believe that our enhanced CCM approach should be able to help us design structures that could potentially scan the entire crystallographic space."
In terms of improving the current generic algorithm, Kumarsays that the current CCM has two continuous variables that are optimized for desired crystal structures. "However," he points out, "with our planned CCM enhancements, this search space will become even larger, and as mentioned replete with local minima that could lead to ambiguous results. We plan to use hybrid genetic, algorithms that use GA in combination with traditional nonlinear optimization approaches to locate the global minima in the large search space of experimental parameters."
Moving forward, Kumar says that other innovations you might develop apart from having ssDNA grafted nanoparticles of spherical shape, the team's collaborators from Brookhaven National Laboratory (and co-authors of the current work) have performed experiments introducing external motifs of different shapes and its effect on final crystal structure. "Different shapes will allow us to control the interaction range and size between particles, and thereby form crystal lattices that are currently inaccessible from using spherical particles. We're already investigating these experiments and developing strategies that could be used in the design of various nanocrystal lattices," he explains.
"Our current research focuses on developing entropy-based models to uniquely define the crystal lattices that would explain the formation of DNA grafted nanomaterials," Kumar continues. "Experiments will validate this model, which will then be used in rational design with the GA framework. In addition, we're planning to extend this rational design strategy to crystal structures with external motifs that aid in self-assembly."
Addressing other areas of research that might benefit from their study, Venkatasubramanian says that the scientists need a new paradigm that increases the idea flow, broadens the search horizon, and archives the knowledge from today's successes to accelerate those of tomorrow. "Our framework addresses this challenge, and in a sense, we're leveraging how nature discovers new materials through the Darwinian model of evolution by suitably marrying it with computational methods. It's Darwin on steroids! This approach can potentially revolutionize materials design," he concludes, "impacting a broad range of products that affect our daily lives, from drugs and agricultural chemicals such as pesticides or herbicides to fuel additives, paints and varnishes, and even personal care products such as shampoo."
More information: Designing DNA-grafted particles that self-assemble into desired crystalline structures using the genetic algorithm, PNAS November 12, 2013 vol. 110 no. 46 18431-18435, doi:10.1073/pnas.1316533110
Related:
1Nanoparticle Superlattice Engineering with DNA, Science 14 October 2011: Vol. 334 no. 6053 pp. 204-208, doi:10.1126/science.1210493
Journal information: Proceedings of the National Academy of Sciences , Science
© 2013 Phys.org. All rights reserved.