How a 'mistake' in a single-cell organism is actually a rewrite essential to life
A tiny but unexpected change to a segment of RNA in a single-cell organism looks a lot like a mistake, but is instead a change to the genetic information that is essential to the organism's survival.
Scientists have discovered this RNA "edit" in Trypanosoma brucei, a parasite that causes sleeping sickness in Africa and Chagas disease in Latin America. Though the organism is a model system for this work, the finding could lead to a new drug target to fight the parasite if higher species don't share this genetic behavior.
Some of the organism's genetic activity was already known. In the case of gene products called tRNAs, which help assemble the amino acids that make proteins, T. brucei was known to have only one tRNA with a specific segment of RNA that ensures the tRNA's proper function. Additionally, examples of RNA editing have been discovered before.
But in this case, the way genetic information necessary for the protein production process was changed – through a swap of three nucleotides for three others that are completely out of place – has never been seen before.
"These are changes for which no chemistry is known and has never been described. We don't know what enzyme is involved and that is the million-dollar question: What mechanism is doing this? We haven't a clue," said Juan Alfonzo, professor of microbiology at The Ohio State University and senior author of the study.
"If the activity is unique to a trypanosome, then you have a good drug target. If it is widespread, then you have to reconsider one more time what coding sequences really mean in the sense that you can indeed change them in a very programmed fashion by activities that don't exist – that have not been described," said Alfonzo, also an investigator in Ohio State's Center for RNA Biology.
The work is the result of Alfonzo's longtime collaboration with co-lead author Christopher Trotta, senior director of biology at PTC Therapeutics in South Plainfield, N.J.
The study appears online in the journal Molecular Cell and is scheduled for print publication on Oct. 24.
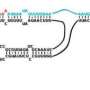
The finding was not only unexpected, but serendipitous. Alfonzo's lab was analyzing an enzyme affecting T. brucei's tRNA behavior in response to a request from Trotta, a drug developer who is considered a pioneer of research on tRNAs. To begin the analysis, Alfonzo sought to identify the intron, a specific segment of RNA, that needs to be removed before the tRNA can participate in the selection of the right amino acids during protein production.
This critical function of removing the intron is called splicing – in essence, a pre-requisite chemical reaction affirming that tRNA can deliver the correct instructions for protein production. If a tRNA is not spliced, it will not work in protein production and the cell will die.
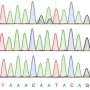
The trouble was, Alfonzo couldn't locate the intron that he knew was there. After multiple attempts, he found that the intron's sequence in this organism changed after transcription, the point at which a copy of RNA is made from a DNA sequence as the first step of gene expression.
This edit – hard to find because of its odd nature – consisted of a change to three nucleotides, the molecules that form DNA and RNA. Because of its rarity and unusual nature, it is called a noncanonical edit.
"It's noncanonical because it is not typical. It is completely not typical," Alfonzo said. "And for the first time, we show the biological significance. We show that if you don't edit, you don't splice. This editing is required for splicing, and splicing is required for functionality. Otherwise, cells die."
Previously known methods of RNA editing include deamination, the removal of sections of molecules from the RNA that change the message from the DNA, and nucleotide insertion, deletion or exchange. The editing described here is a swap of three nucleotides for three others that, according to the rules of biology, do not belong where they end up. This is why it looks like a mistake.
Colleagues have suggested that this edit should have been identified by researchers who do deep sequencing, which involves repeated readings of all nucleotides within an RNA molecule, Alfonzo noted. But he is not surprised that technology didn't yield these results.
"In massive sequencing, you match RNAs to the sequence in the genome. Any mismatch is called a sequence mistake and is thrown in the trash. So this noncanonical editing may well be in the trash bin of many of these deep sequencing researchers," he said.
Journal information: Molecular Cell
Provided by The Ohio State University