Researchers develop first-of-its-kind mathematical model for biological process that keeps immune system working
Scientists at USC have created a mathematical model that explains and predicts the biological process that creates antibody diversity – the phenomenon that keeps us healthy by generating robust immune systems through hypermutation.
The work is a collaboration between Myron Goodman, professor of biological sciences and chemistry at the USC Dornsife College of Letters, Arts and Sciences; and Chi Mak, professor of chemistry at USC Dornsife.
"To me, it was the holy grail," Goodman said. "We can now predict the motion of a key enzyme that initiates hypermutations in immunoglobulin (Ig) genes."
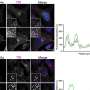
Goodman first described the process that creates antibody diversity two years ago. In short, an enzyme called "activation-induced deoxycytidine deaminase" (or AID) moves up and down single-stranded DNA that encodes the pattern for antibodies and sporadically alters the strand by converting one nitrogen base to another, which is called "deamination." The change creates DNA with a different pattern – a mutation.
These mutations, which AID creates a million-fold times more often than would otherwise occur, generate antibodies of all different sorts – giving you protection against germs that your body hasn't even seen yet.
"It's why when I sneeze, you don't die," Goodman said.
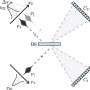
In studying the seemingly random motion of AID up and down DNA, Goodman wanted to understand why it moved how it did, and why it deaminated in some places much more than others.
"We looked at the raw data and asked what the enzyme was doing to create that," Goodman said. He and his team were able to develop statistical models whose probabilities roughly matched the data well, and were even able to trace individual enzymes visually and watch them work. But they were all just approximations, albeit reasonable ones.
Collaborating with Mak, however, offered something better: a rigorous mathematical model that describes the enzyme's motion and interaction with the DNA and an algorithm for directly reading out AID's dynamics from the mutation patterns.
At the time, Mak was working on the mathematics of quantum mechanics. Using similar techniques, Mak was able to help generate the model, which has been shown through testing to be accurate.
"Mathematics is the universal language behind physical science, but its central role in interpreting biology is just beginning to be recognized," Mak said. Goodman and Mak collaborated on the research with Phuong Pham, assistant research professor, and Samir Afif, a graduate student at USC Dornsife. An article on their work, which will appear in print in the Journal of Biological Chemistry on October 11, was selected by the journal as a "paper of the week."
Next, the team will generalize the mathematical model to study the "real life" action of AID as it initiates mutations during the transcription of Ig variable and constant regions, which is the process needed to generate immunodiversity in human B-cells.
Journal information: Journal of Biological Chemistry
Provided by University of Southern California