A tighter fit with artificial DNA
An artificial base that enhances the protein-binding affinity and selectivity of DNA expands the DNA machinery.DNA aptamers are expected to play an important role in the diagnosis and treatment of various cancers and other gene-related diseases. These nucleic acids, which bind target substances such as proteins and cells, are typically generated through in vitro evolution methods. Ichiro Hirao and colleagues from the RIKEN Center for Life Science Technologies have now developed a new generation strategy based on an expanded genetic alphabet that improves the affinity and selectivity of DNA aptamers1.
The generation of DNA aptamers by in vitro evolution involves several rounds of selection and amplification using a library of nucleic acid sequences until a target-specific strand is produced. Unfortunately, the isolated strand often exhibits inadequate affinity and selectivity, and modification of the natural bases or sugars of the nucleotides can result in the production of new nucleic acids without greatly enhancing the abilities of the DNA aptamers themselves.
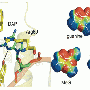
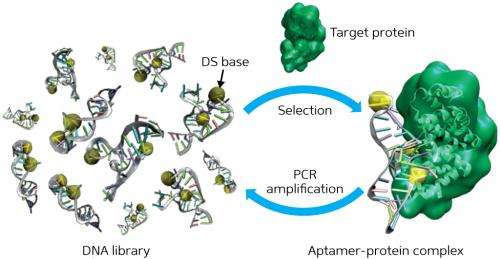
Postulating that increasing the number of bases might improve the binding of DNA aptamers, Hirao and his colleagues incorporated an unnatural base, Ds, into a nucleic acid library in addition to the four natural DNA bases—adenine, guanine, thymine and cytosine.
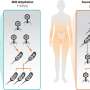
To create their library, the researchers synthesized nucleic acids that contained Ds bases at only a few predetermined positions in randomized natural base sequences. After exposing the library to the target substance, they isolated the high-affinity strands and amplified them by polymerase chain reaction (PCR) for additional selection rounds (Fig. 1). They repeated the selection–amplification process while increasing the selection requirements until they obtained the desired aptamers.
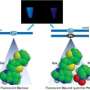
Hirao's team found that the newly generated Ds-containing aptamers showed significantly higher affinity for two human target proteins than known aptamers that only contained natural bases. Moreover, by replacing all the Ds bases with adenine in these new aptamers, the researchers observed that the affinities were reduced by several hundred times, confirming the importance of the Ds bases in improving the binding of a target.
Hirao explains that inclusion of a fifth base might increase the structural diversity of DNA to better fit its target. Furthermore, unlike natural bases, Ds is highly hydrophobic, which might tighten interactions with hydrophobic parts of target proteins.
"Aptamer technology is only one example of the possible applications for our unnatural base pair system in synthetic biology," says Hirao. His team is currently using this system to label functional RNA molecules and generate new proteins with non-standard amino acids. The team is also trying to create artificial cells based on six different DNA bases.
More information: Nature Biotechnology 31, 453–457 (2013). DOI: 10.1038/nbt.2556
Journal information: Nature Biotechnology
Provided by RIKEN