Novel chemistry for new class of antibiotic
University of Adelaide research has produced a potential new antibiotic which could help in the battle against bacterial resistance to antibiotics.
The potential new antibiotic targets a bacterial enzyme critical to metabolic processes.
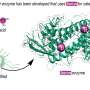
The compound is a protein inhibitor which binds to the enzyme (called biotin protein ligase), stopping its action and interrupting the life cycle of the bacteria.
"Existing antibiotics target the bacterial cell membranes but this potential new antibiotic operates in a completely different way," says Professor Andrew Abell, project leader and Acting Head of the University's School of Chemistry and Physics.
Professor Abell says the compound, although at a very early stage of development - it has not yet been tested on an animal model - has the potential to become the first of a new class of antibiotics.
"Bacteria quickly build resistance against the known classes of antibiotics and this is causing a significant global health problem," he says.
"Preliminary results show that this new class of compound may be effective against a wide range of bacterial diseases, including tuberculosis which has developed a strain resistant to all known antibiotics."
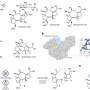
Developing the new protein inhibitor involved a novel approach called 'in situ click chemistry'. A selection of small molecules, or 'precursor fragments', are presented to the bacteria in a way so that the target protein enzyme itself builds the inhibiting compound and also binds with it.
"In a sense the bacteria unwittingly chooses a compound that will stop its growth and assembles it - like building a weapon and using it against itself," says Professor Abell. "We've gone a step further to specifically engineer the enzyme so that it builds the best and most potent weapon."
"Our results are promising. We've made the compounds; we know they bind and inhibit this enzyme and we've shown they stop the growth of a range of bacteria in the laboratory. The next critical step will be investigating their efficacy in an animal model."
"Thanks to this new approach what might have taken a year or more with a range of sequential experiments, we can now do in one single experiment," Professor Abell says.
The research has been published in the journal Chemical Science and is in collaboration with researchers at Monash University and Adelaide's Women's and Children's Hospital.
More information: pubs.rsc.org/en/content/articl … g/2013/sc/c3sc51127h
Journal information: Chemical Science
Provided by University of Adelaide