New technique allows biologists to profile patterns of gene regulation in rare cell populations
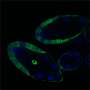
Mapping all of the chemical, or epigenetic, changes to chromosomes that affect which genes are turned on or off—and thus determine the fate of genomically identical cells in the body—usually requires a large amount of starting cellular material. This technical limitation has impeded the analysis of gene regulation in many rare cell types and in small clinical biopsy samples. Now, a team of biologists led by A*STAR scientists has developed a protocol for characterizing these changes that requires up to 100 times fewer cells than previously needed. As a proof of principle, the researchers used the approach to chart which genes are activated or repressed in mouse reproductive cells that eventually give rise to eggs or sperm.
"We developed a technique for epigenetic analysis of very small numbers of cells, and used it to identify gene regulatory elements in the genome of germ cells in the mouse embryo," says one of the team leaders, Shyam Prabhakar from A*STAR's Genome Institute of Singapore.
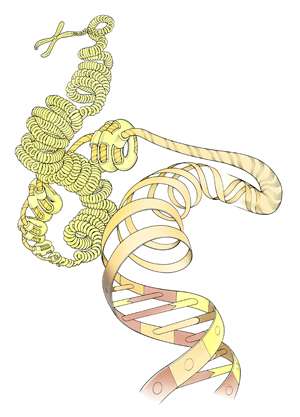
The team, which is co-led by the Genome Institute of Singapore's Huck Hui Ng, started with a standard technique for epigenetic analysis known as ChIP-seq. This method combines a way of investigating the interactions between proteins and DNA called 'chromatin immunoprecipitation', or ChIP, with high-throughput DNA sequencing. ChIP-seq can reveal modifications to chromosome structures that affect gene expression (see image). Typically, ChIP-seq requires millions of cells to work properly. Prabhakar and Ng's team, however, adapted the technique so that it could work with just 10,000 cells. This involved miniaturizing various aspects of the protocol so that it worked reliably with smaller volumes.
Using the small-scale ChIP-seq method on germ cells taken from embryonic mice, the researchers revealed many previously unknown epigenetic features needed for proper maintenance and development of these early precursors of eggs and sperm. They include changes to promoters and enhancer elements that control how germ-cell specific genes are activated, as well as repressive marks on some genes that are needed only later in development. Interestingly, the analysis also showed that genetic elements called retrotransposons and genes involved in activating immune responses are both silenced, preserving genomic integrity and preventing cell death in these important reproductive cells.
Next, Prabhakar, Ng and co-workers plan to apply the method to study rare cell populations in humans, including biopsies taken from cancer patients. "This technology now enables us to study other cell types of limited quantity," Ng says.
More information: Ng, J.-H., Kumar, V., Muratani, M., Kraus, P., Yeo, J.-C. et al. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Developmental Cell 24, 324–333 (2013). www.cell.com/developmental-cel … ii/S1534580712005850
Journal information: Developmental Cell