A trick to fold proteins more quickly
A team of researchers of the International School for Advanced Studies (SISSA) of Trieste and of University of Cambridge have devised a method to reduce the time used to simulate how proteins take on their signature three-dimensional shape. Such important information to comprehend their function is usually obtained using often very costly experimental techniques.
To understand how proteins work it is important to know their three-dimensional shape, but also the way it is produced. We need to know, in other words, how the amino acid filament which makes up the proteins is capable of folding over itself to take on a specific shape.
Today the study of molecular dynamics of proteins is based on computer simulations in which the system is treated as a three-dimensional set of balls (1 ball = 1 atom) observed while it evolves through time. This is a very accurate but rather slow technique, therefore a group of researchers, including Daniele Granata and Alessandro Laio at SISSA of Trieste, have devised a trick to reduce simulation times.
"We exploit the experimental data obtained observing the proteins through nuclear magnetic resonance, and use them to create restraints to be applied to the model", explained Laio, who has coordinated the research published in Proceedings of the National Academy of Sciences (PNAS).
"Basically, we used a 'trick'. Imagine I pulled your arm and directed you towards a certain place, let's say, to have a reference, from Trieste to Rimini. The trick enables to reach a destination in a period of time even a thousand times inferior to what usually required for that itinerary. Thanks to mathematical rules– gathered from previous observations carried out on 'trips to Rimini' – I can calculate, starting from the fixed travel time, how long it would have taken the same individual to spontaneously reach the same destination without being pulled. The same assumption may be applied to a protein that must fold in order to take on a certain shape."
"There is a law that connects the time a protein takes to fold using the 'trick' and the time used to do it spontaneously," explains Laio.
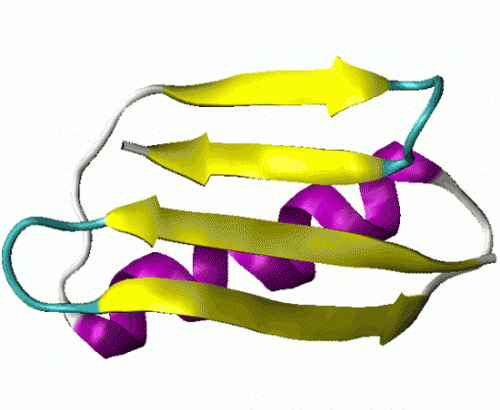
The technique employed by Granata and Laio, applied in this case to streptococcal protein G, has enabled to obtain results which are perfectly consistent with those obtained using the more common technique, with the advantage it remarkably reduces calculation times. "Considering the slowness of standard computer molecular dynamics techniques, methods like ours optimize processing times and could give a big boost to research."
More information: Granata, D. et al. Characterization of the free-energy landscapes of proteins by NMR-guided metadynamics, PNAS, April 23, 2013, vol. 110, no. 17, 6817–6822. www.pnas.org/cgi/doi/10.1073/pnas.1218350110
Journal information: Proceedings of the National Academy of Sciences
Provided by Sissa Medialab