Do-it-yourself viruses: How viruses self assemble
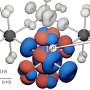
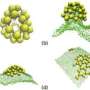
A new model of the how the protein coat (capsid) of viruses assembles, published in BioMed Central's open access journal BMC Biophysics, shows that the construction of intermediate structures prior to final capsid production (hierarchical assembly) can be more efficient than constructing the capsid protein by protein (direct assembly). The capsid enveloping a virus is essential for protection and propagation of the viral genome. Many viruses have evolved a self-assembly method which is so successful that the viral capsid can self assemble even when removed from its host cell.
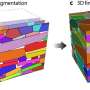
The construction of large protein structures has been observed experimentally but the mechanism behind this is not well understood. Even the 'simple' icosahedral protein coat of the T1 virus requires integration of 60 protein components. Computational models of the physical interactions of component proteins are used to investigate the dynamics and physical constraints that regulate whether the components assemble correctly.
Using computer simulations a team from the Institute for Theoretical Physics and the Center for Quantitative Biology (BioQuant), University of Heidelberg, has compared direct and hierarchical assembly methods for T1 and T3 viruses. The team led by Ulrich S Schwarz, realized that direct assembly often led to the formation of unfavorable intermediates, especially when the dissociation rate was low, which hindered further assembly, causing the process to stall. In contrast, for many conditions hierarchical assembly was more reliable, especially if the bonds involved had a low dissociation rate.
Discussing the practical applications of these results, Dr Schwarz commented, "Hierarchical assembly has not been systematically investigated before. Theoretical models and computer simulations, like ours, can be used to understand the mechanism behind assembly of complex viruses and give an indication of how other large protein complexes assemble."
He continued, " With our computer simulations, we are now in a position to investigate systems which are too large to be studied by molecular resolution. This rational approach might have many applications not only in biomedicine, but also in materials science, where many researchers strive to learn from nature how to assembly complex structures."
More information: Stochastic Dynamics of Virus Capsid Formation: Direct versus Hierarchical Self-Assembly Johanna E Baschek, Heinrich CR Klein and Ulrich S Schwarz, BMC Biophysics (Section: Computational and theoretical biophysics) (in press)
Provided by BioMed Central